# fit regular mediationlibrary(JSmediation)mediation_fit <-mdt_simple(med1,IV =group,DV = emobias,M = L_amyg_post)add_index(mediation_fit)
Test of mediation (simple mediation)
==============================================
Variables:
- IV: group
- DV: emobias
- M: L_amyg_post
Paths:
==== ============== ===== =======================
Path Point estimate SE APA
==== ============== ===== =======================
a -0.087 0.030 t(145) = 2.86, p = .005
b 1.466 0.621 t(140) = 2.36, p = .020
c -0.458 0.224 t(141) = 2.05, p = .042
c' -0.313 0.228 t(140) = 1.37, p = .173
==== ============== ===== =======================
Indirect effect index:
- type: Indirect effect
- point estimate: -0.127
- confidence interval:
- method: Monte Carlo (5000 iterations)
- level: 0.05
- CI: [-0.294; -0.0125]
Fitted models:
- X -> Y
- X -> M
- X + M -> Y
The mdt_simple package does not allow for covars. You might get a sig total path if you do not include the pre-intervention measure. This differs from the paper. Below is how you can include the covariate.
Here we are fitting a mediation model with lavaan (controlling for pre-intervention scores)
Code
library(lavaan)sem1 <-'#b and cp and covaremobias ~ b * L_amyg_post + cp * group + covar*L_amyg_pre#aL_amyg_post ~ a * group# indirect 1indirect1 := a * b# totaltotal := cp + (a * b) #prob mediated#prop_indirect1prop_med_1 := indirect1 / (indirect1+cp)'fit <-sem(model = sem1, data = med1, se ="bootstrap", bootstrap =5000)# you should run 5000-10000 bootstraps
Mediation analysis was carried out using the lavaan package in R. I tested whether left amygdala-mPFC connectivity post-intervention mediates the association between Condition and Positive Emotional Memory Bias. I controlled for the effects of pre-intervention amygdala-mPFC connectivity on the mediator and outcome variable. 10,000 bootstrap samples were used. I replicated the patterns of significance found by Cho et al. (2023). Effect estimates slightly varied from the original study, which may be due to the use of bootstrapping which involves random sampling.
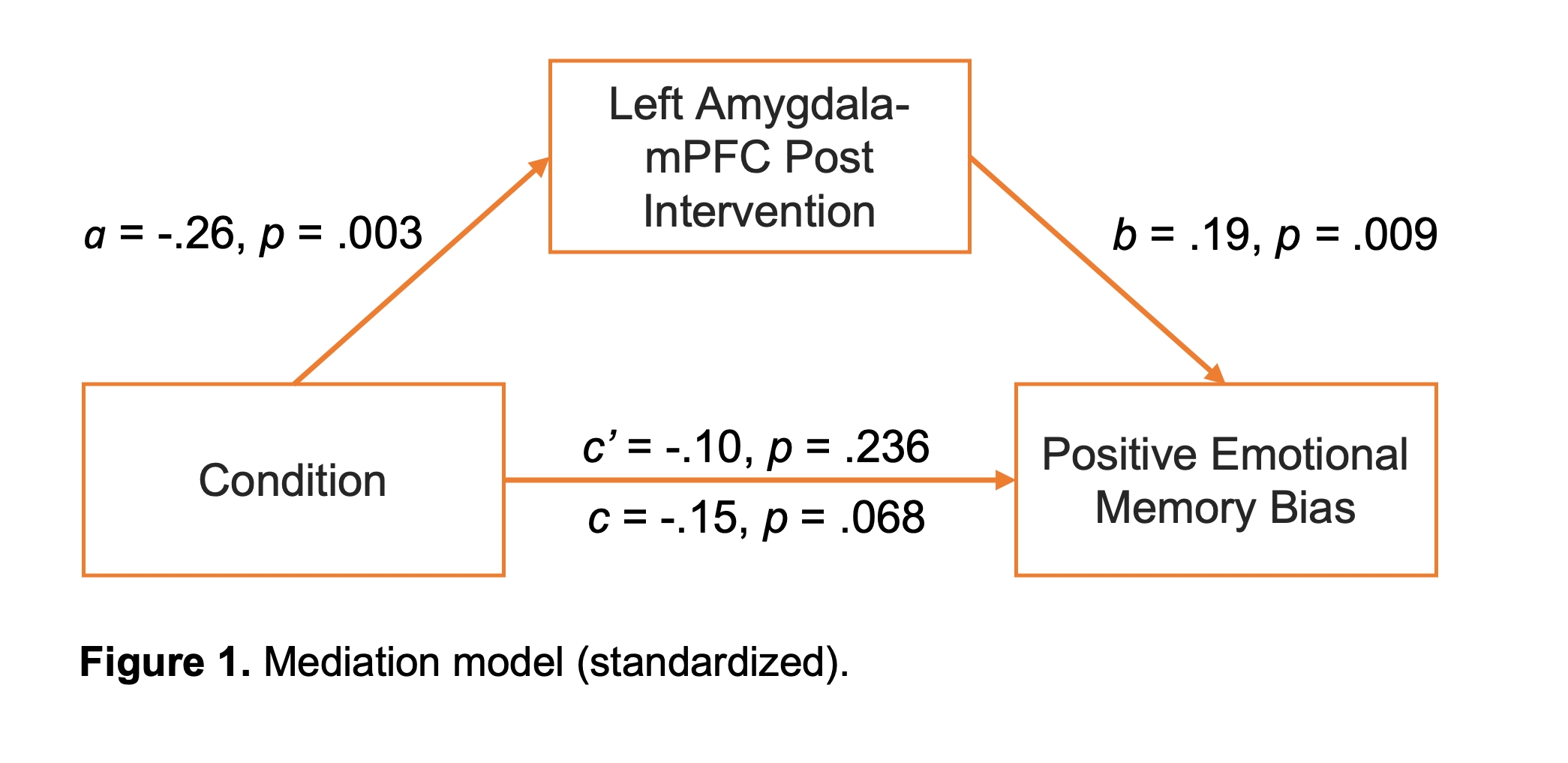
In the present model (Figure 1), the bootstrapped indirect effect was significant, ab = -0.15, p = 0.02, 95% CI [-0.29,-0.029], standardized effect = -0.049. The direct effect was not significant, c’ = -0.279, p = 0.236, 95% CI [-0.756, 0.178] , standardized effect = -0.104. The total effect was not significant, c = -0.412, p = 0.068, 95% CI [-0.858, 0.028], standardized effect = -0.153. Thus, according to the causal steps approach, there was no mediation (the total path was no significant). However, according to the joint significance test and the boostrapped test, there was a significant mediation effect. Thus, the results indicate that post-intervention amygdala-mPFC connectivity mediated the association between Condition and bias scores. I really appreciate that the authors shared their data openly on OSF and OpenNeuro. However, the authors indicated in the paper that analysis material was also available, but I wasn’t able to find this. This would have been helpful to confirm how they specified their mediation model.
Source Code
---title: "Lab 08: Mediation Answers"subtitle: "Lab 8"date: last-modifiedformat: html: self-contained: true anchor-sections: true code-tools: true code-fold: true fig-width: 8 fig-height: 4 mainfont: Source Sans Pro theme: journal toc: true toc-depth: 3 toc-location: left captions: true cap-location: margin table-captions: true tbl-cap-location: margin reference-location: marginexecute: message: false warning: false---## Load DataYou can read in data from osf directly.```{r}library(tidyverse)library(MeMoBootR)library(lavaan)library(easystats)library(knitr)url <-"https://osf.io/4yfwq//?action=download"filename <-"Data.txt"data <-read.delim(url, header =TRUE, sep ="\t",dec =".")```## Tidy- Clean up the dataset from OSF```{r}med1 <- data %>%filter(mPFC.LAmyg_post !="N/A", emobiasscore !="N/A") %>%mutate(L_amyg_post=as.numeric(mPFC.LAmyg_post), emobias =as.numeric(emobiasscore), group=Group..1.inc..2.dec., L_amyg_pre=as.numeric(`mPFC.LAmyg_pre`)) %>%select(group, emobias, L_amyg_post, L_amyg_pre) %>%mutate(group=ifelse(group==1, -0.5, 0.5))```## Simple mediation```{r}# fit regular mediationlibrary(JSmediation)mediation_fit <-mdt_simple(med1,IV =group,DV = emobias,M = L_amyg_post)add_index(mediation_fit)```- The `mdt_simple` package does not allow for covars. You might get a sig total path if you do not include the pre-intervention measure. This differs from the paper. Below is how you can include the covariate.## Simple mediation with covariate```{r}#| eval: false#| library(MeMoBootR)med1 <-mediation1(x =group,y = emobias,m = L_amyg_pos, cvs=c("L_amyg_pre"))```## LavaanHere we are fitting a mediation model with lavaan (controlling for pre-intervention scores)```{r}library(lavaan)sem1 <-'#b and cp and covaremobias ~ b * L_amyg_post + cp * group + covar*L_amyg_pre#aL_amyg_post ~ a * group# indirect 1indirect1 := a * b# totaltotal := cp + (a * b) #prob mediated#prop_indirect1prop_med_1 := indirect1 / (indirect1+cp)'fit <-sem(model = sem1, data = med1, se ="bootstrap", bootstrap =5000)# you should run 5000-10000 bootstraps``````{r}lavaan::parameterestimates(fit, ci=TRUE, boot.ci.type ="perc") %>%kable()```## Write-upMediation analysis was carried out using the *lavaan* package in R. I tested whether left amygdala-mPFC connectivity post-intervention mediates the association between Condition and Positive Emotional Memory Bias. I controlled for the effects of pre-intervention amygdala-mPFC connectivity on the mediator and outcome variable. 10,000 bootstrap samples were used. I replicated the patterns of significance found by Cho et al. (2023). Effect estimates slightly varied from the original study, which may be due to the use of bootstrapping which involves random sampling.In the present model (Figure 1), the bootstrapped indirect effect was significant, ab = -0.15, *p* = 0.02, 95% CI [-0.29,-0.029], standardized effect = -0.049. The direct effect was not significant, *c'* = -0.279, *p* = 0.236, 95% CI [-0.756, 0.178] , standardized effect = -0.104. The total effect was not significant, *c* = -0.412, *p* = 0.068, 95% CI [-0.858, 0.028], standardized effect = -0.153. Thus, according to the causal steps approach, there was no mediation (the total path was no significant). However, according to the joint significance test and the boostrapped test, there was a significant mediation effect. Thus, the results indicate that post-intervention amygdala-mPFC connectivity mediated the association between Condition and bias scores. I really appreciate that the authors shared their data openly on OSF and OpenNeuro. However, the authors indicated in the paper that analysis material was also available, but I wasn't able to find this. This would have been helpful to confirm how they specified their mediation model.