Generalized Linear Madness: Binary Outcome Models/Logistic Regression
Princeton University
2024-02-19
Packages
- Access the .qmd document here: https://github.com/jgeller112/PSY504-Advanced-Stats-S24/blob/main/slides/04-logistic/logisticqmd.qmd
Today
Nuts and bolts of logistic regression
- Binomial/Bernoulli distribution
- Logit link
- Log-odds, odds, probabilities
- Likelihood
Motivating example: Weight loss, gender, BMI
- Modeling fitting
- Parameter interpretation
- Model fit and model diagnostics
- Comparing models
- Visualizing
- Reporting
Linear model
Everything so far has included a continuous outcome
Model \(y\) as a linear function of predictors
\[ Y_i = b_0 + b_1*x + e_i\]
\[Y \sim~Normal(\mu, \sigma)\]
Dichotomous outcomes
The simplest kind of categorical data is each case is binary
- Correct/incorrect
- Pass/fail
- Choose/Don’t choose
- Support/Not support
- Relapse/Not relapse
Note
We usually code these as 0s and 1s and model the probability of 1 occurring
Bye bye linear model

Linear models are not appropriate for this type of data
Makes impossible predictions (values not between 0, 1)
Non-normality of residuals
- \(\hat{p}\) or (1-\(\hat{p}\))
Heteroskedasticity
- Variance is influenced by \(\hat{p}\)
Generalized linear model
Regression models with non-normal response likelihoods
\[\begin{align*} Y_i & \sim \mathrm{Dist}(\mu_i, \tau) \\ g(\mu_i) & = \eta_i \\ \eta_i & = \beta_0 + \beta_1 X_{1i} + \beta_2 X_{2i} + \ldots \end{align*}\]Where:
\(y\) is referenced with \(\mu\)
\(g\) is link function (in logistic regression we use logit link)
What distribution do we use to model this process?
We assume that there is some probability p of responding correctly (or selecting what is coded as 1)
We don’t know this probability, so we want to use the data to estimate it
Bernoulli distribution
Distribution of a single, discrete binary outcome \[y\sim Bernoulli(p)\]
A Bernoulli distribution generates a 1 (“success”) with probability p
And a 0 (“failure”) with probability 1−p=q
- \(\mu\) = E(X)=p
- \(\sigma^2(X)=p(1-p)\)
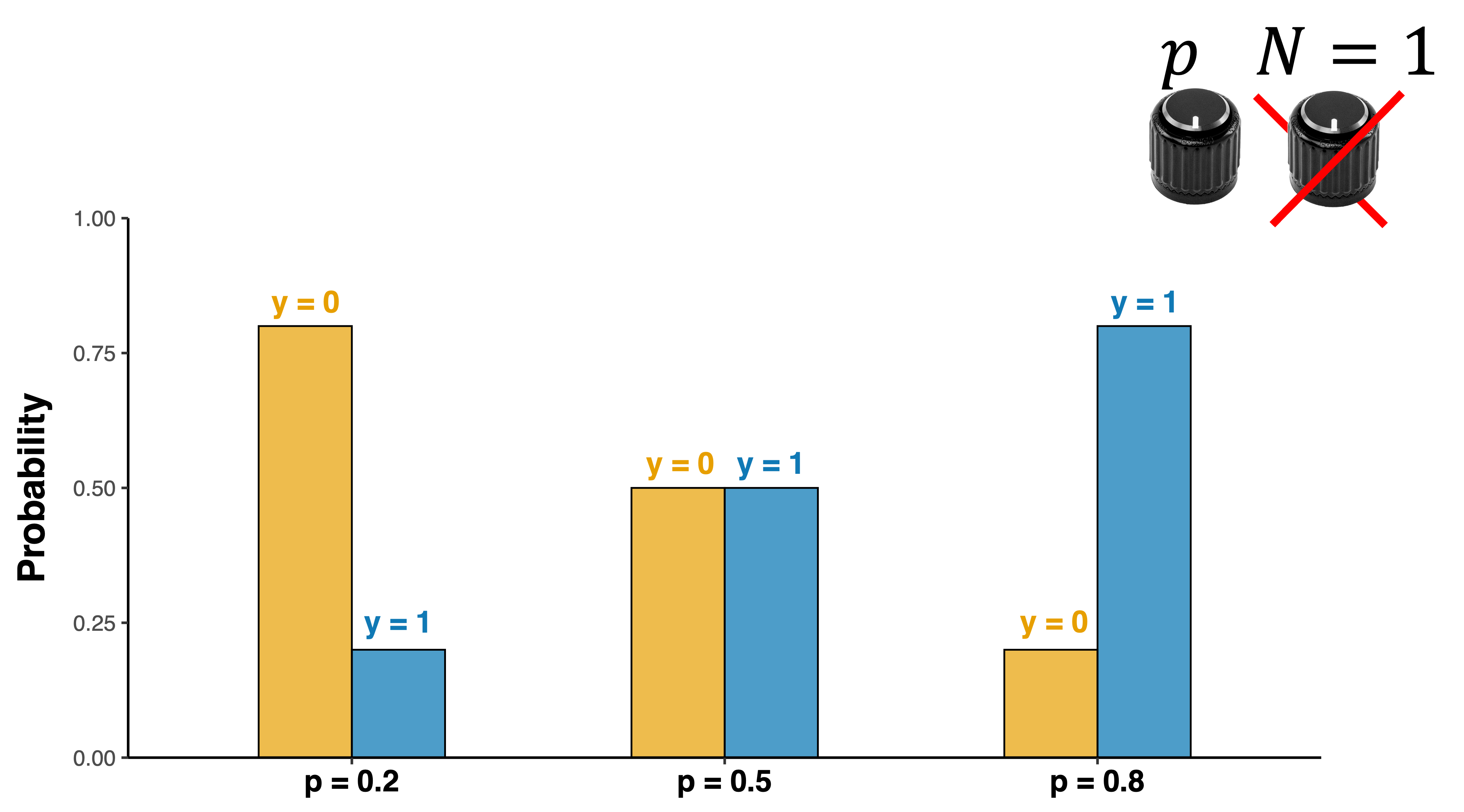
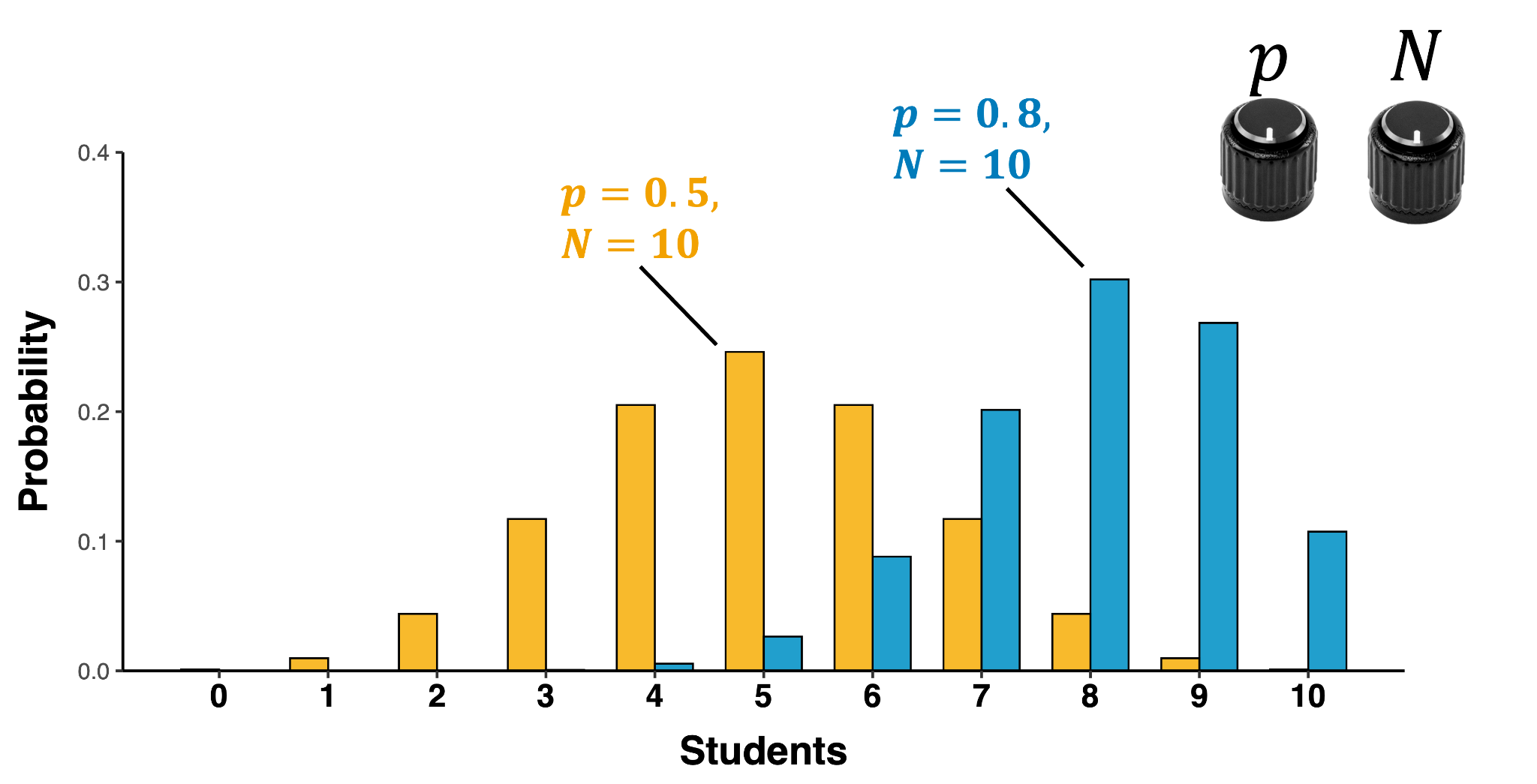
Binomial distribution
Distribution of discrete counts of independent outcomes over a certain set of trials
\[y\sim binomial(N,p)\]
- N = number of trials
- p = probability of “y = 1”
- \(\mu\) = E(X)=np$
- \(\sigma^2(X)=np(1-p)\)
- \(\sigma(X)\)=\(\sqrt{np(1-p)}\)
Fitting a logistic regression in R
In R we use the
glmfunction as opposed tolmlibrary(fivethirtyeight) bechdel=fivethirtyeight::bechdel #fit <- glm( # data = bechdel, # family = binomial(link="logit"), # binary ~ 1 + budget_2013) bechdel %>% dplyr::select(title, binary) %>% head() %>% kable()title binary 21 & Over FAIL Dredd 3D PASS 12 Years a Slave FAIL 2 Guns FAIL 42 FAIL 47 Ronin FAIL
Logit link
- The logit link transforms a linear model in the log-odds metric:
\[\begin{equation*} \log\left(\frac{p}{1 - p}\right)=b_0+b_1X \end{equation*}\]
- To a non-linear model in the probability metric:
\[\begin{equation*} p(logit)=\frac{e^{logit}}{1+e^{logit}} \end{equation*}=\frac{exp(logit)}{1+exp(logit)}\]
Logit link
Step 1
Transform probability into odds
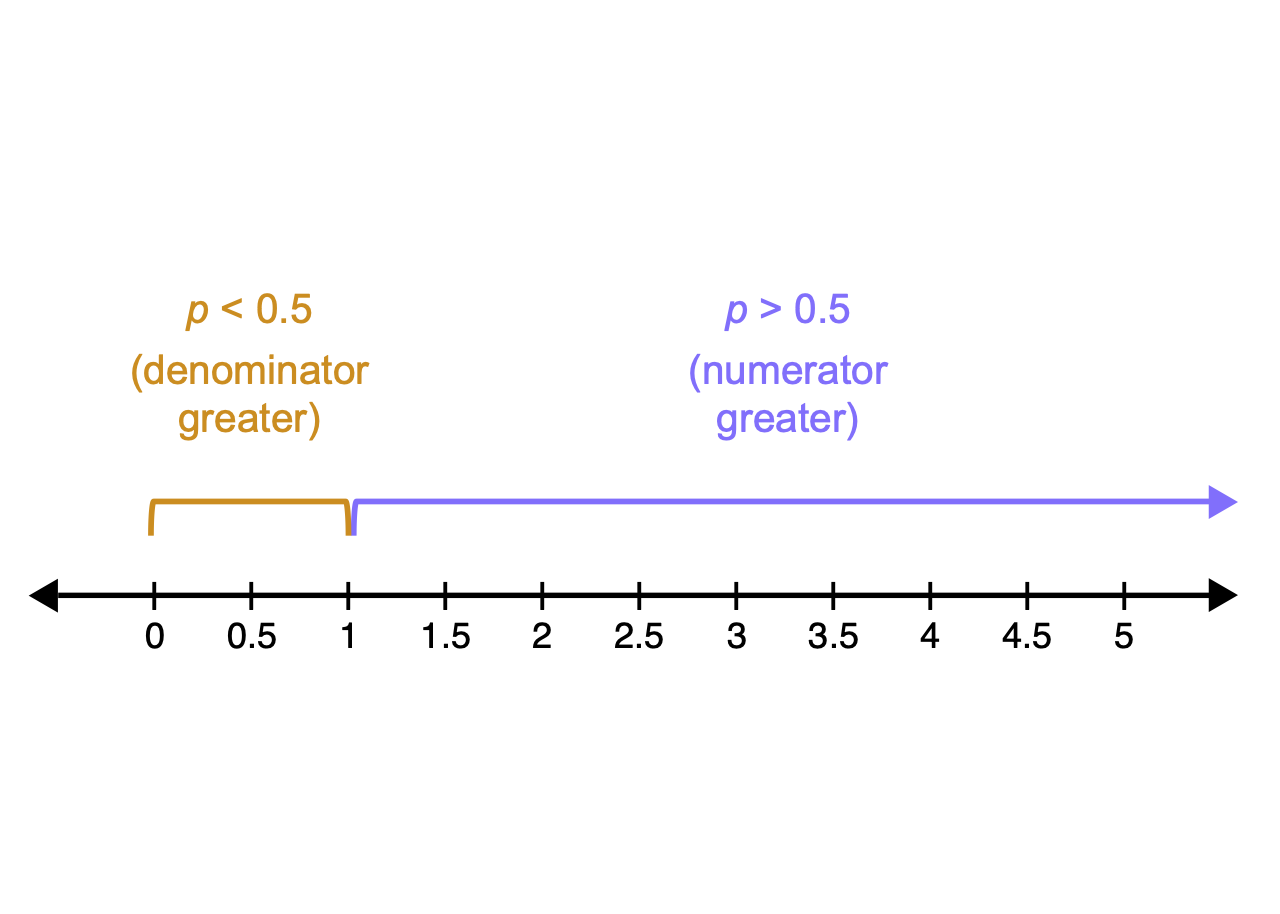
- The “odds” of an event are just the probability it happens divided by the probability it does not happen
\[\textrm{Odds} = \frac{\# \textrm{successes}}{\# \textrm{failures}}= \frac{\# \textrm{successes}/n}{\# \textrm{failures}/n}= \frac{p}{1-p}\]
Logit Link
Step 2
When we convert a probability to odds, odds always > 0
Asymmetric
Curvy
- Problematic for linear model
Solution: take log of the odds
\[logit = log(\frac{p}{1-p})\]
Logit link
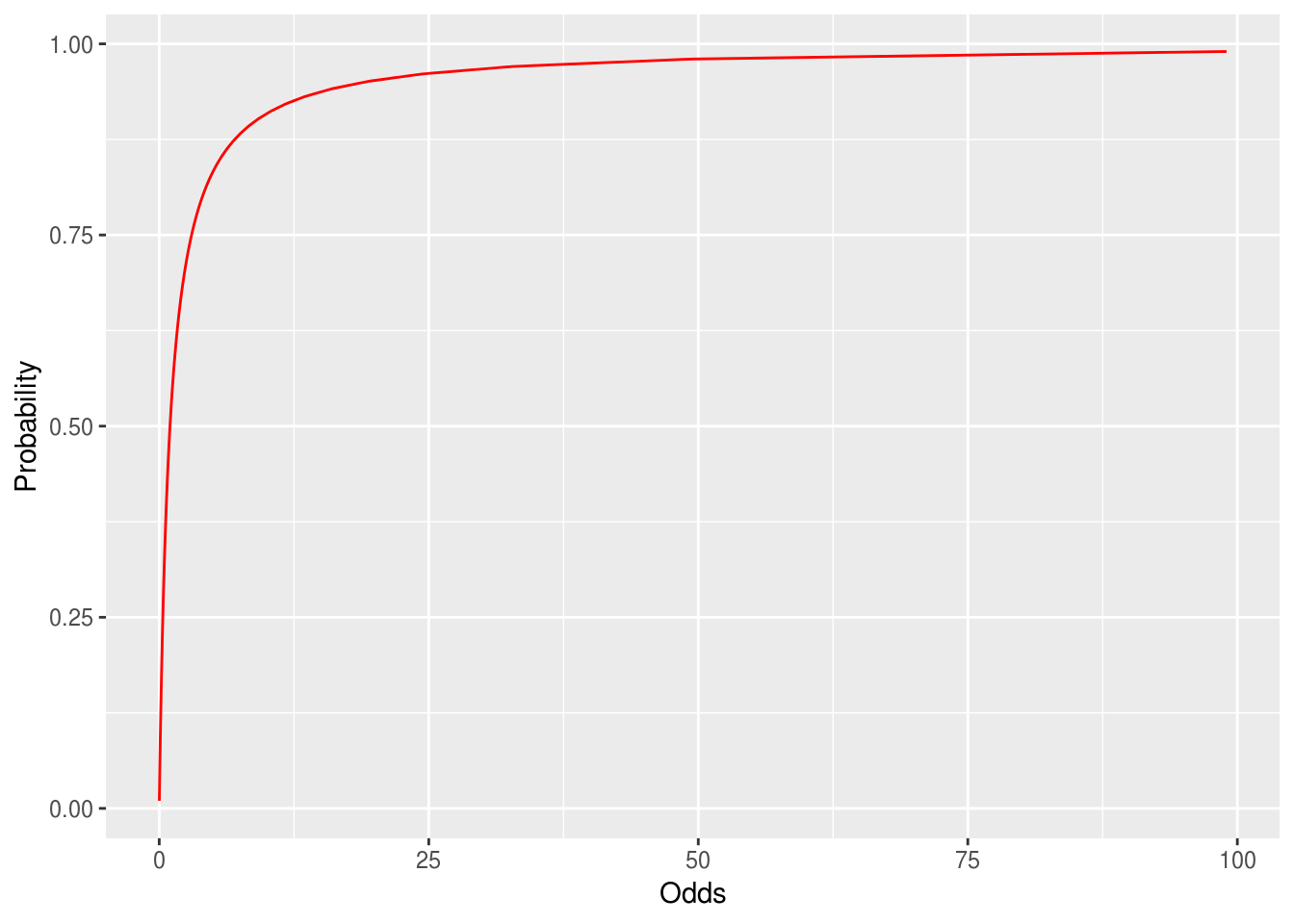
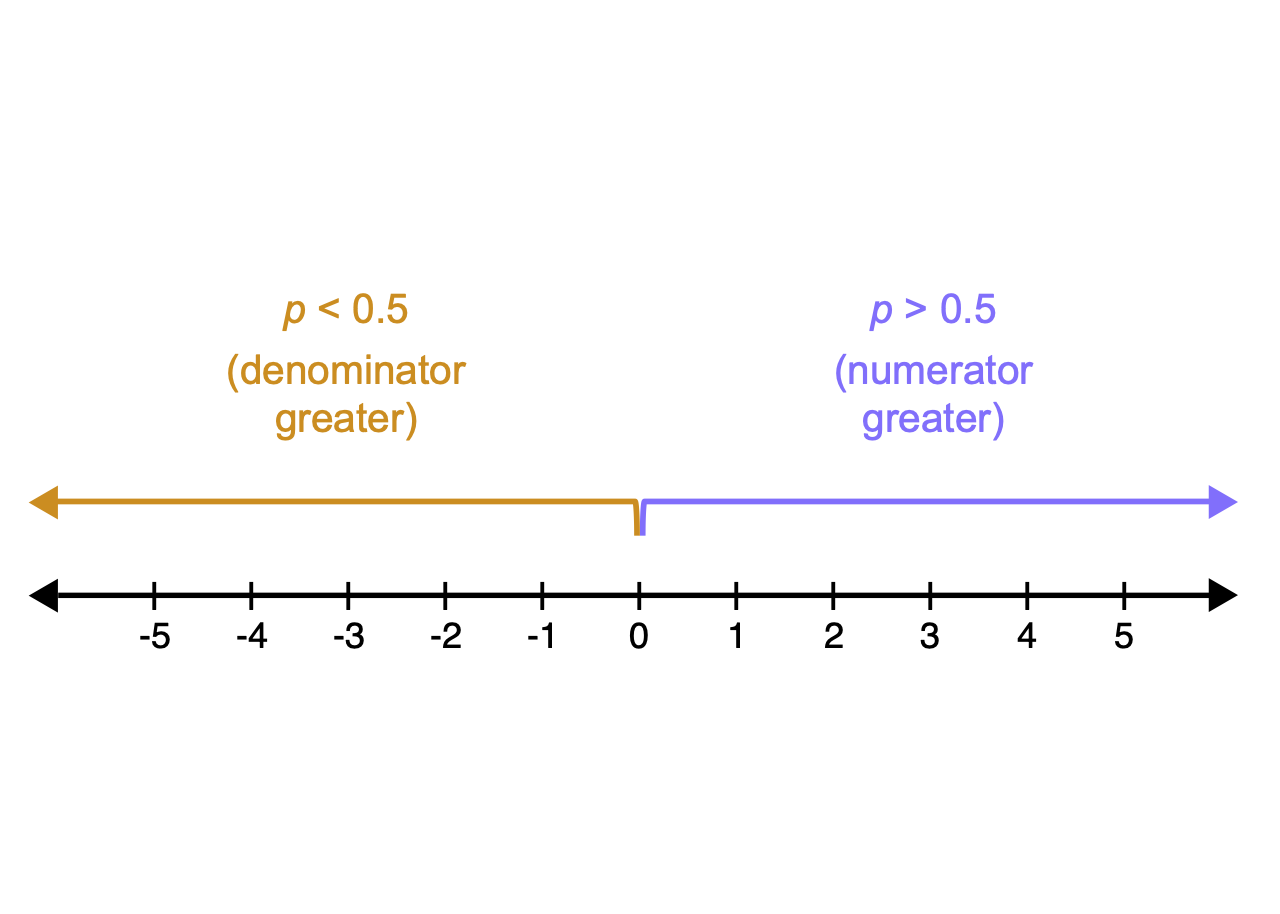
This log odds conversion has a nice property
It converts odds of less than one to negative numbers, because the log of a number between 0 and 1 is always negative
Our data is now linear!
and symmetric
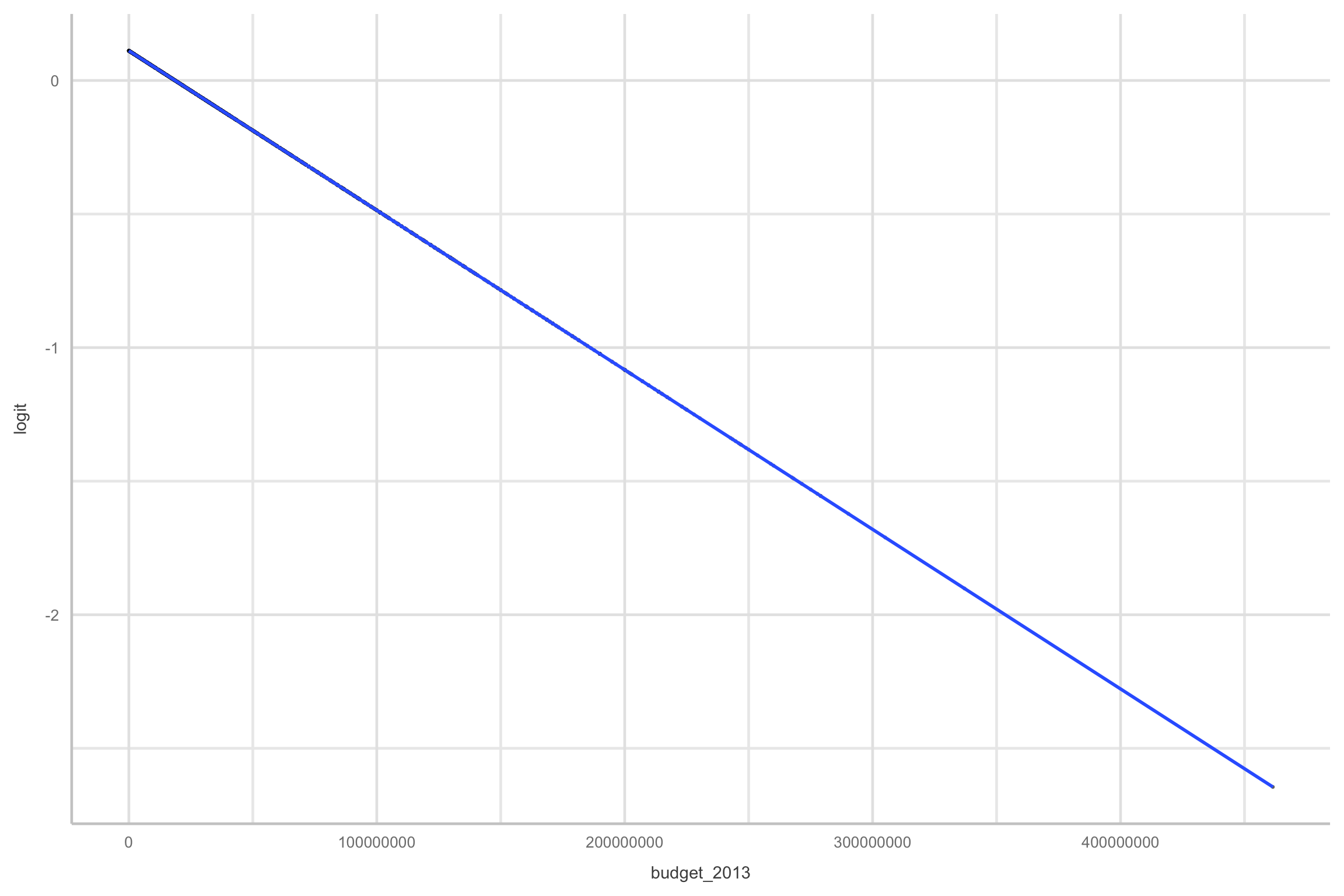
However…
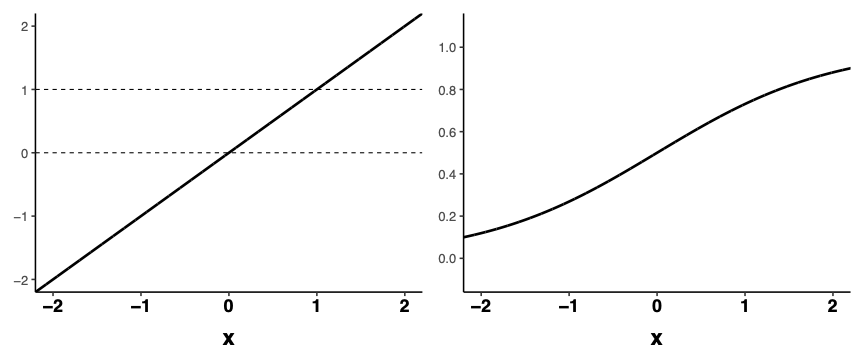
- We need a function that scrunches the output of the linear predictor into a range appropriate for this parameter
Squiggles
We need a function that scrunches the output of the linear predictor into a range appropriate for this parameter
A squiggle (logistic or Sigmoid curve) to the rescue!
Logistic transform
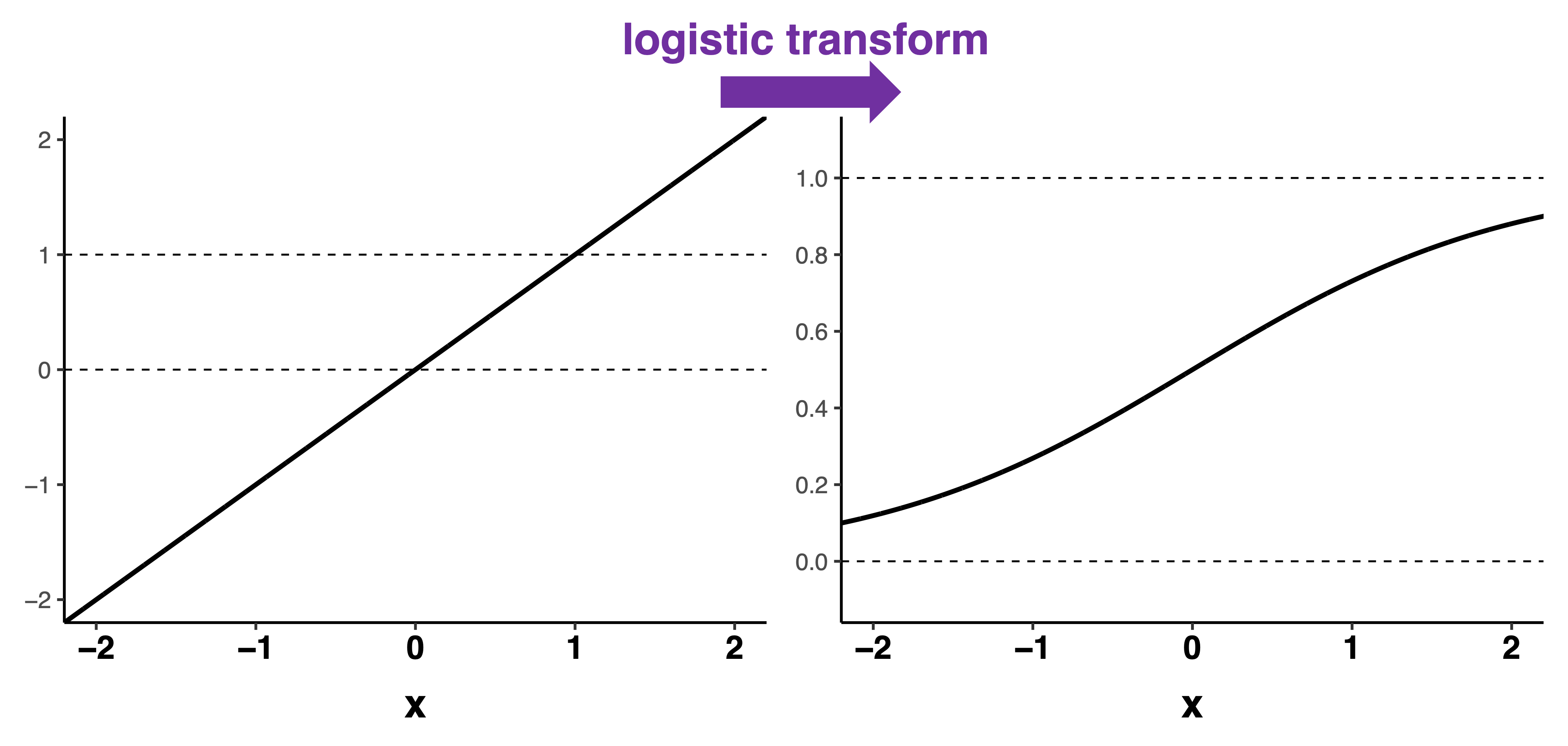
\[p(logit)=\frac{e^{logit}}{1+e^{{logit}}}\]
Slope and intercept parameters
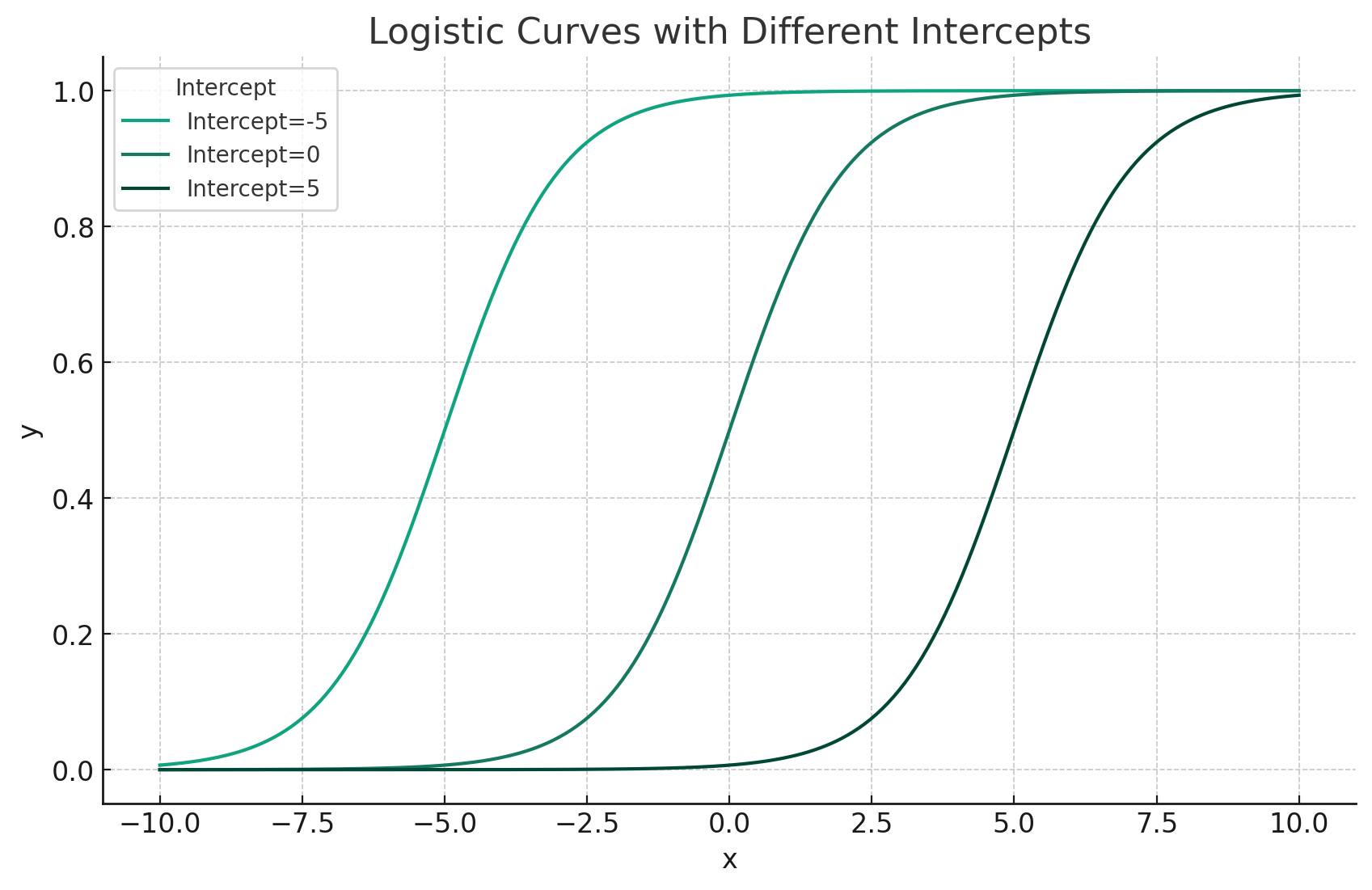
- Intercept moves curve left or right
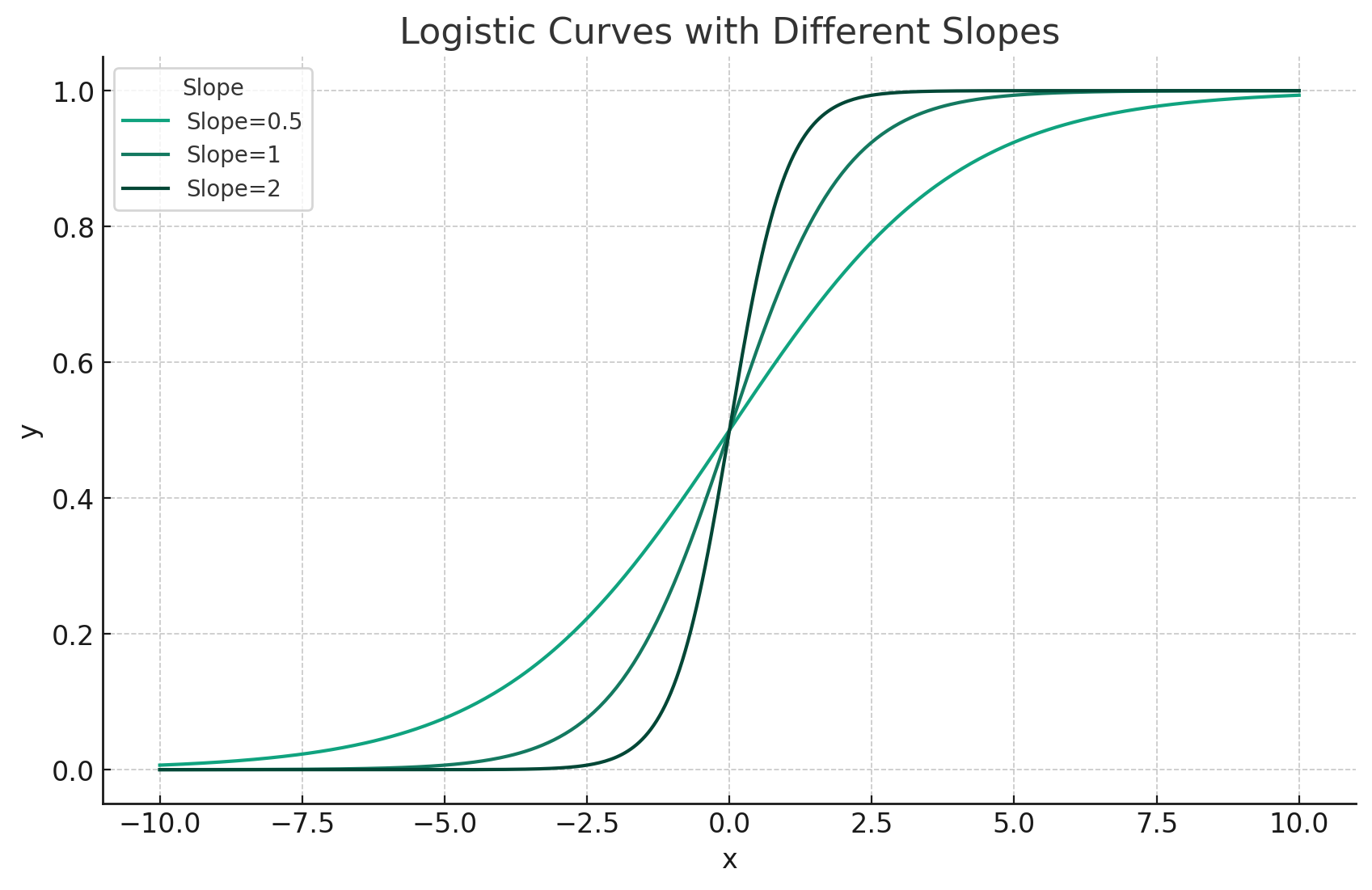
- Slope determines the steepness of the curve
All together
Andrew Heiss
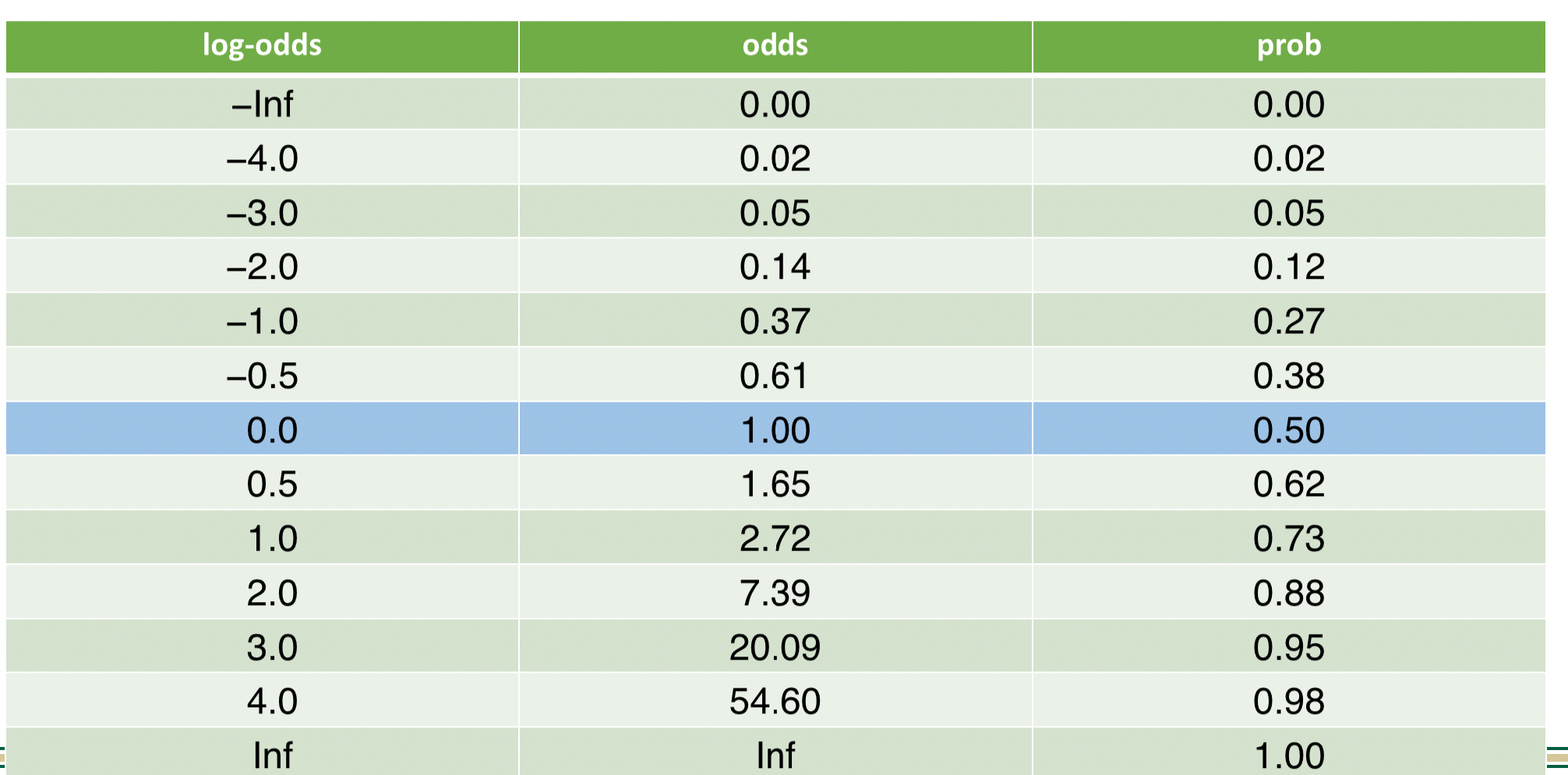
Probabilities, Odds, and Log Odds
Probabilities, Odds, and Log Odds
- From probabilities to log-odds
- From log-odds to probabilities
Probabilities, Odds, and Log Odds
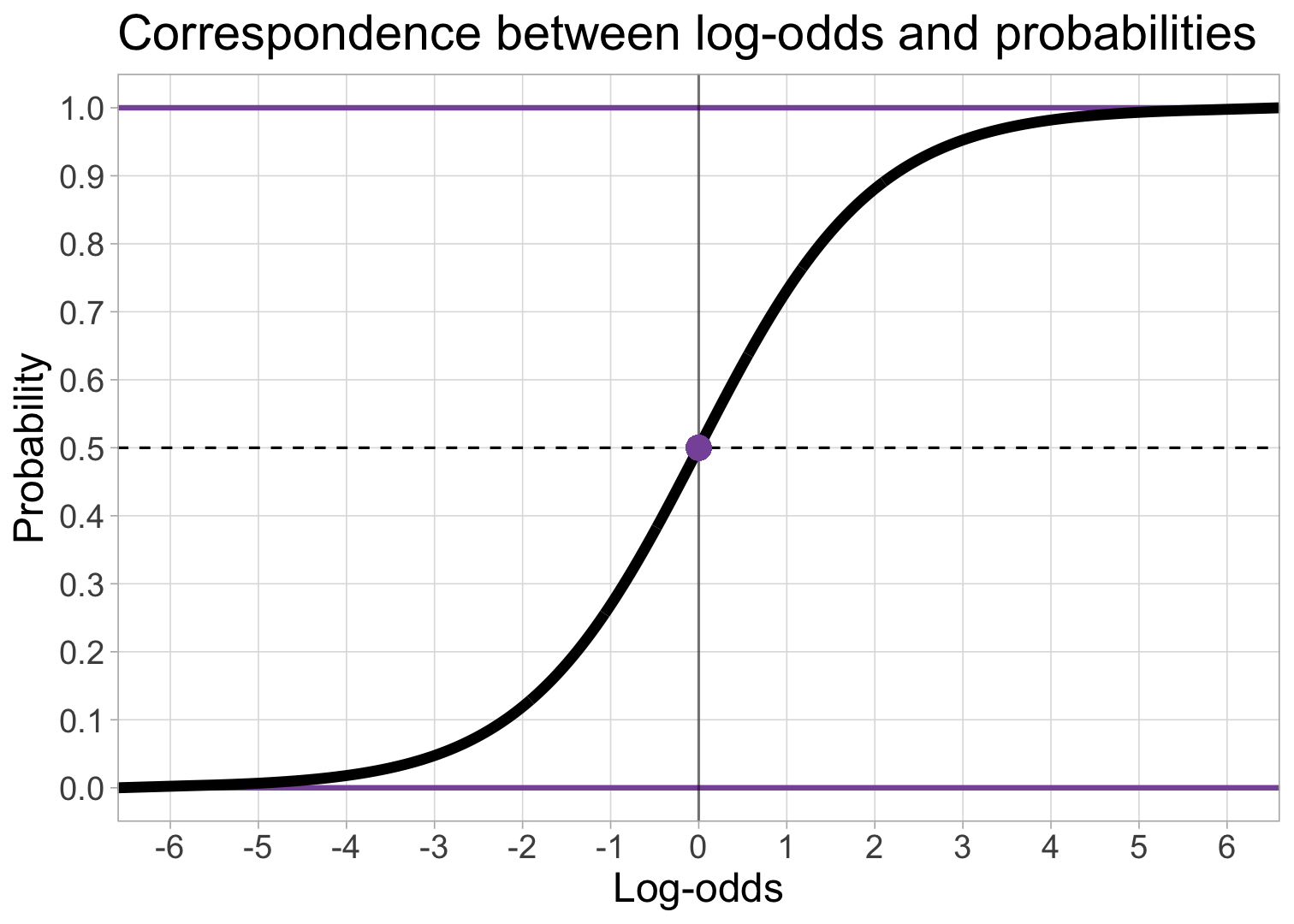
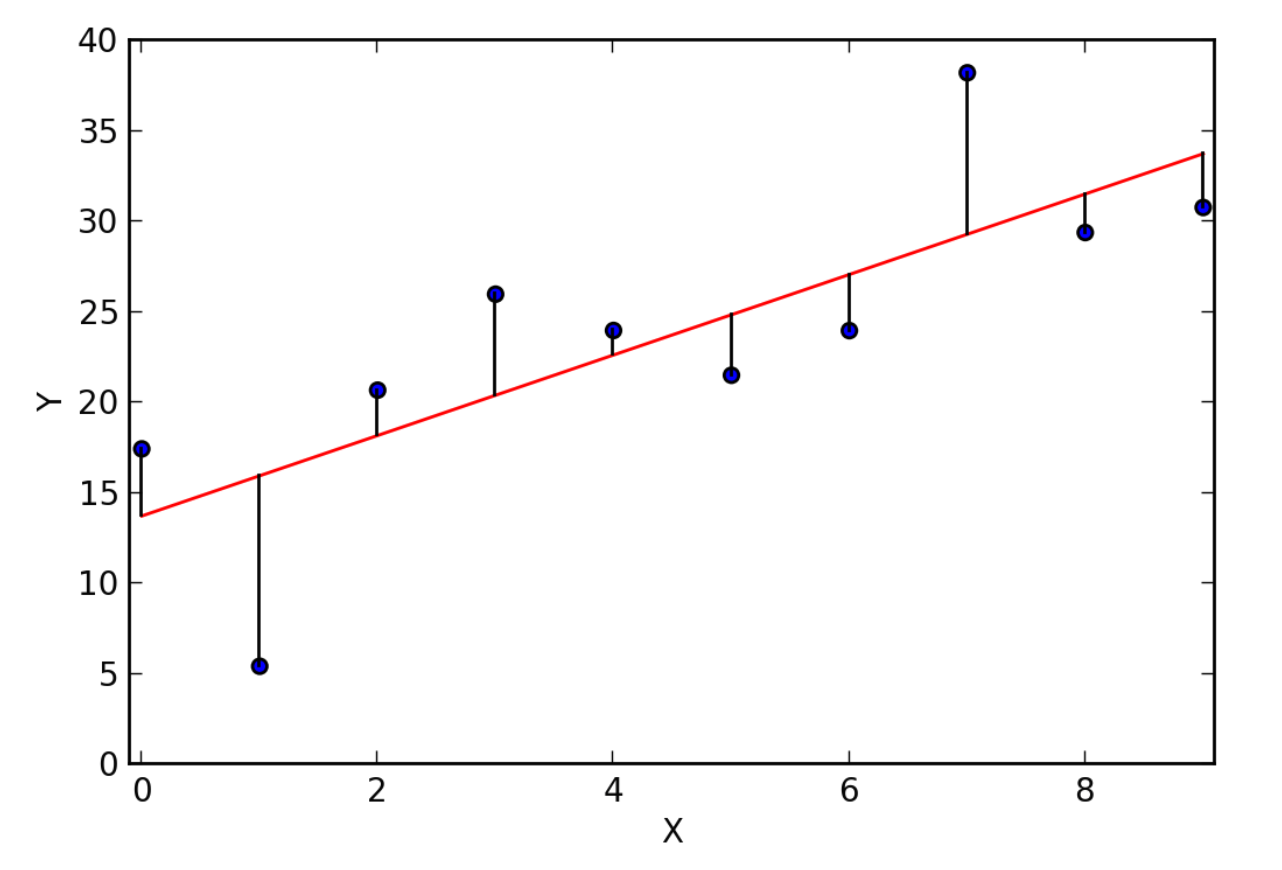
Best fitting squiggle
- In standard linear regression we use least squares to find the best fitting line
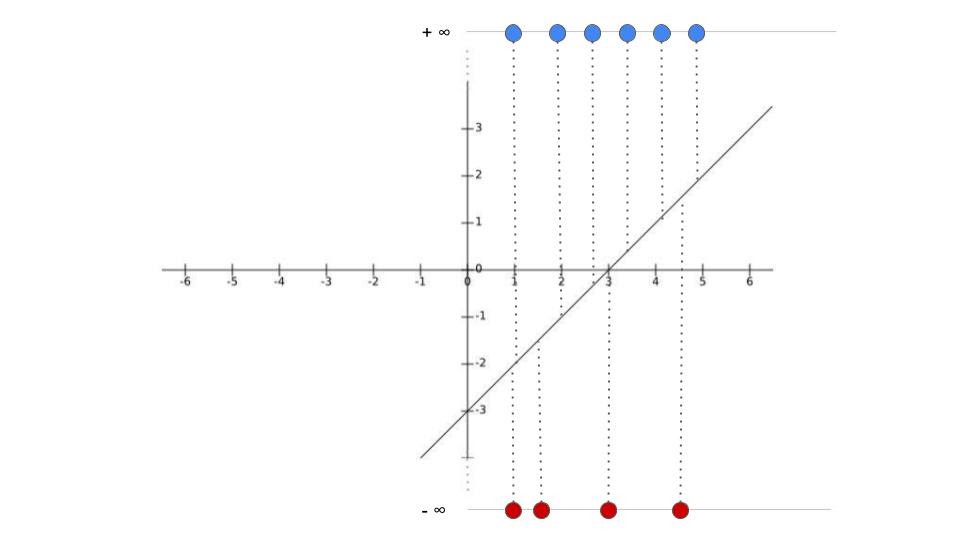
Maximum likelihood estimation
- We need find where points lie on line and get corresponding logits
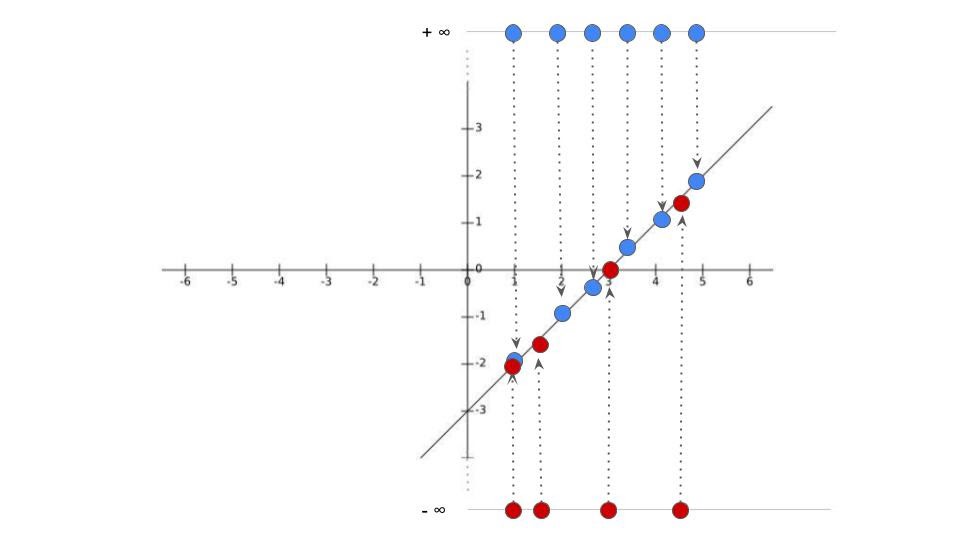
ML: Logistic Regression
- Plot them as a function of probability
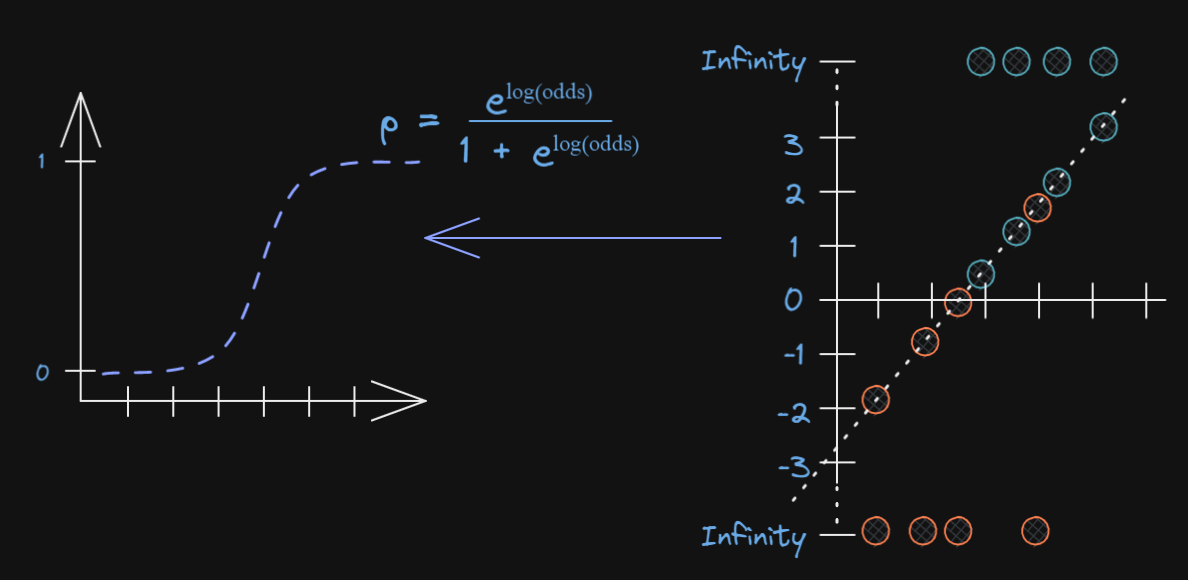
Now what?
We keep rotating the log odds(line) and projecting data on to it and transforming into probabilities
- Until we find maximum likelihood!
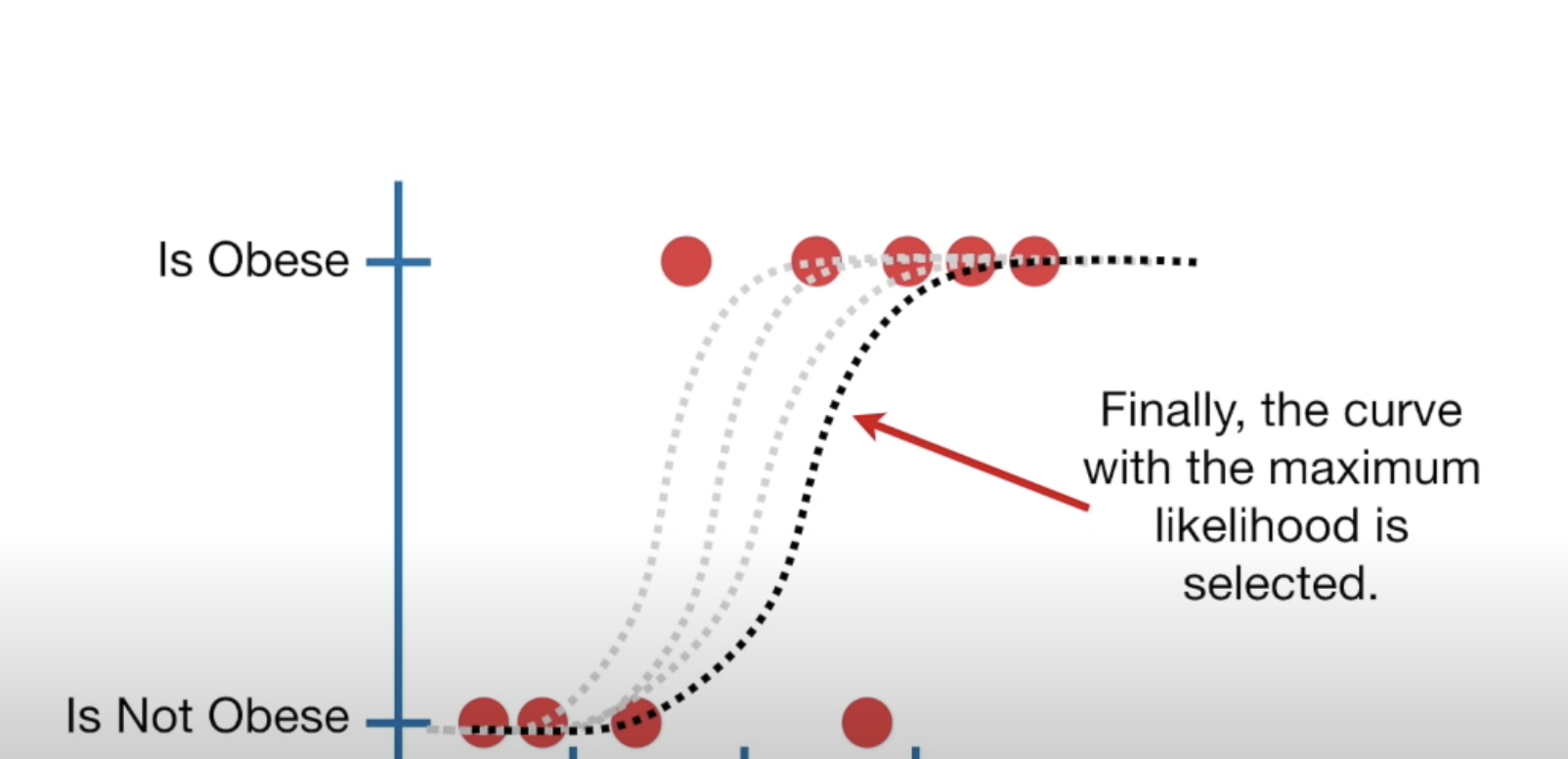
Case Study: Losing Weight
These data are a sample of 500 teens from data collected in 2009 through the U.S. Youth Risk Behavior Surveillance System (YRBSS)
Are the odds that young females report trying to lose weight greater than the odds that males do? Is increasing BMI associated with an interest in losing weight, regardless of sex?
Trying to lose weight
- Selected variables:
DV: *lose.wt.01, which is coded 1 when someone indicated they were trying to lost weight and 0 when they were not trying to. This is a dichotomized version of the lose.wt factor variable.
Our two predictor variables will be:
sexand its related dummy variablefemalebmipct, which is the percentile for a given BMI for members of the same sex
Exploratory data analysis
- Here are two versions of comparing
lose.wt.01bysex
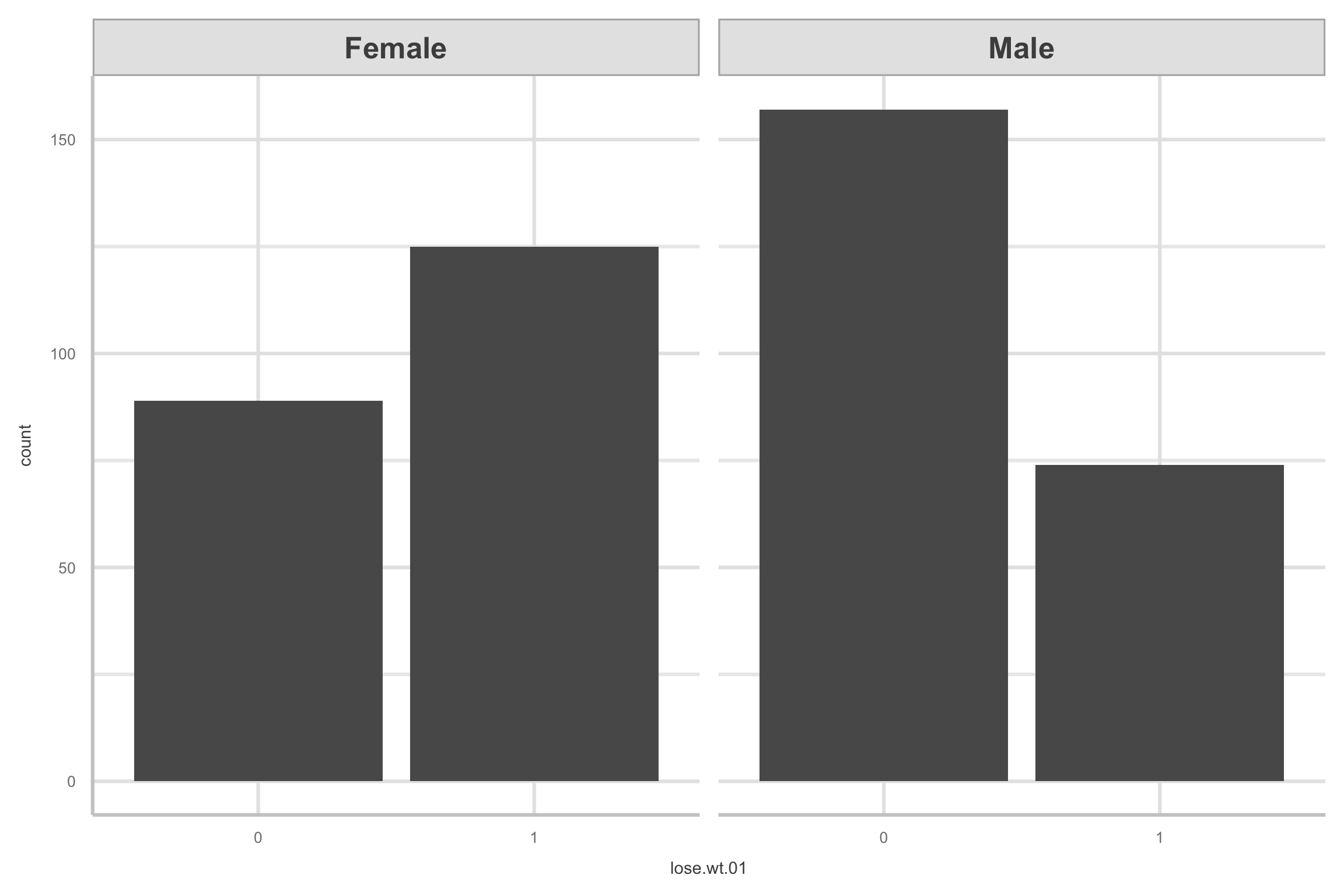
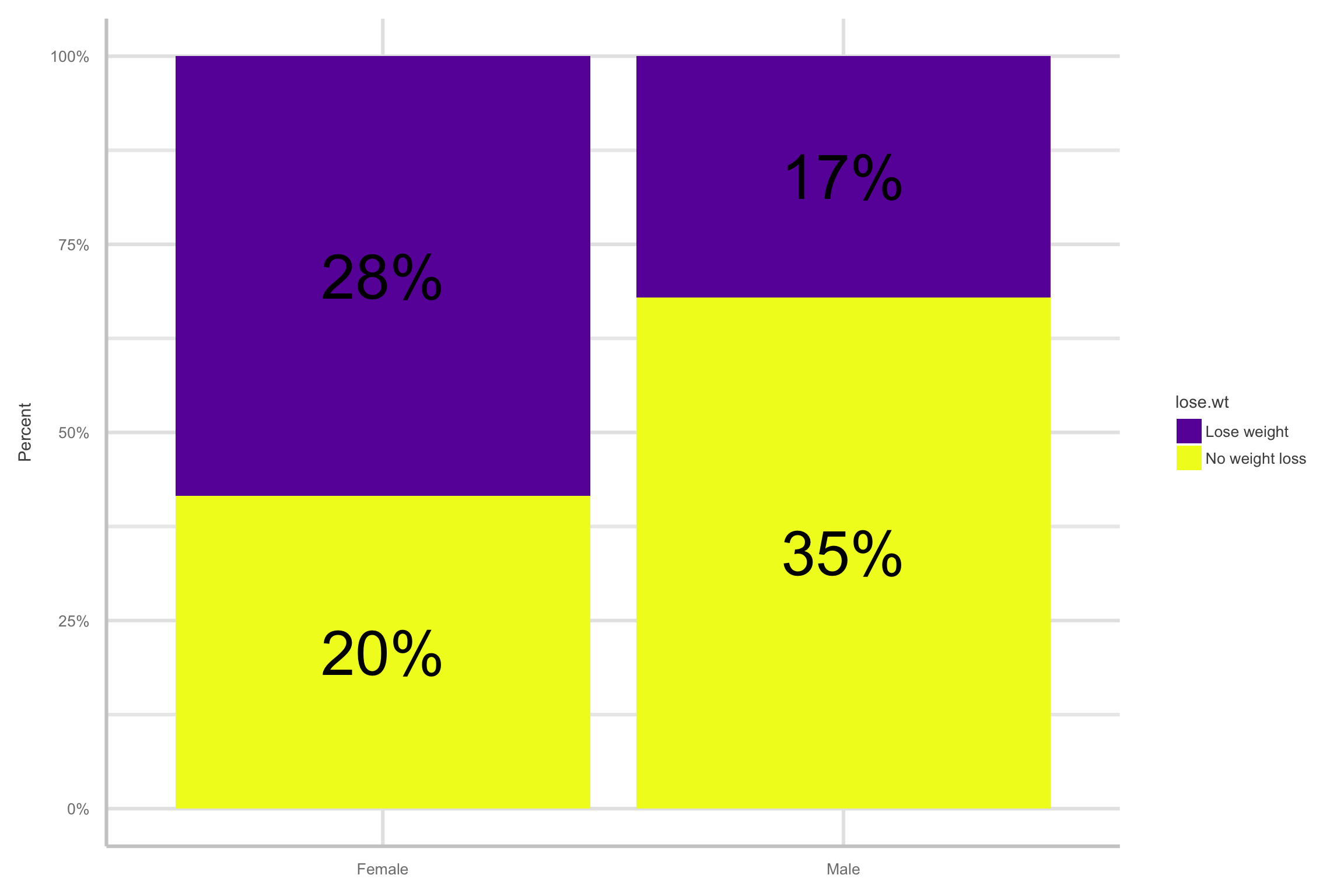
Exploratory data analysis
- Here are the histograms for
bmipct, faceted bylose.wt.
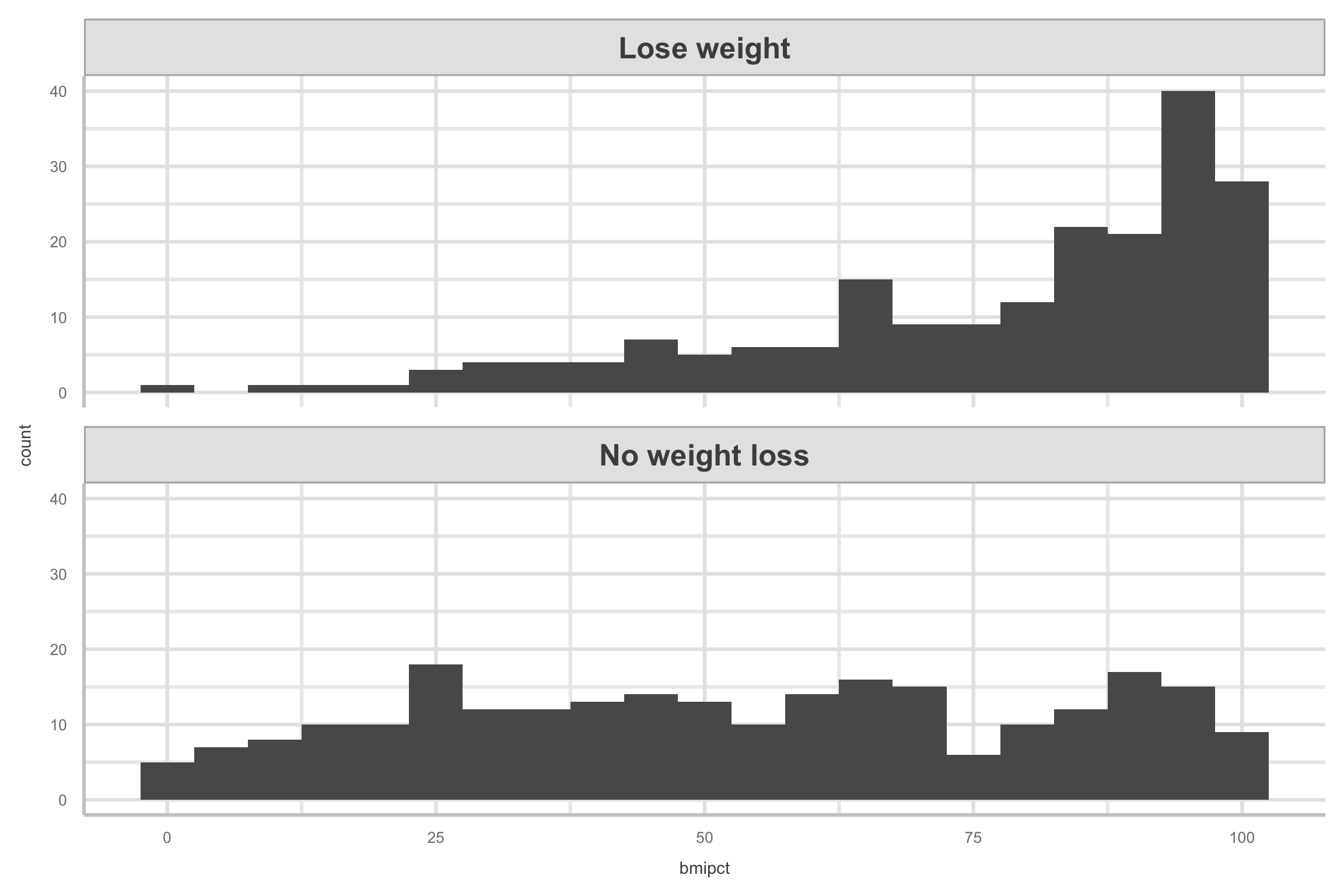
Exploratory Data Analysis
- Here are the histograms for
bmipct, faceted by bothlose.wtandsex
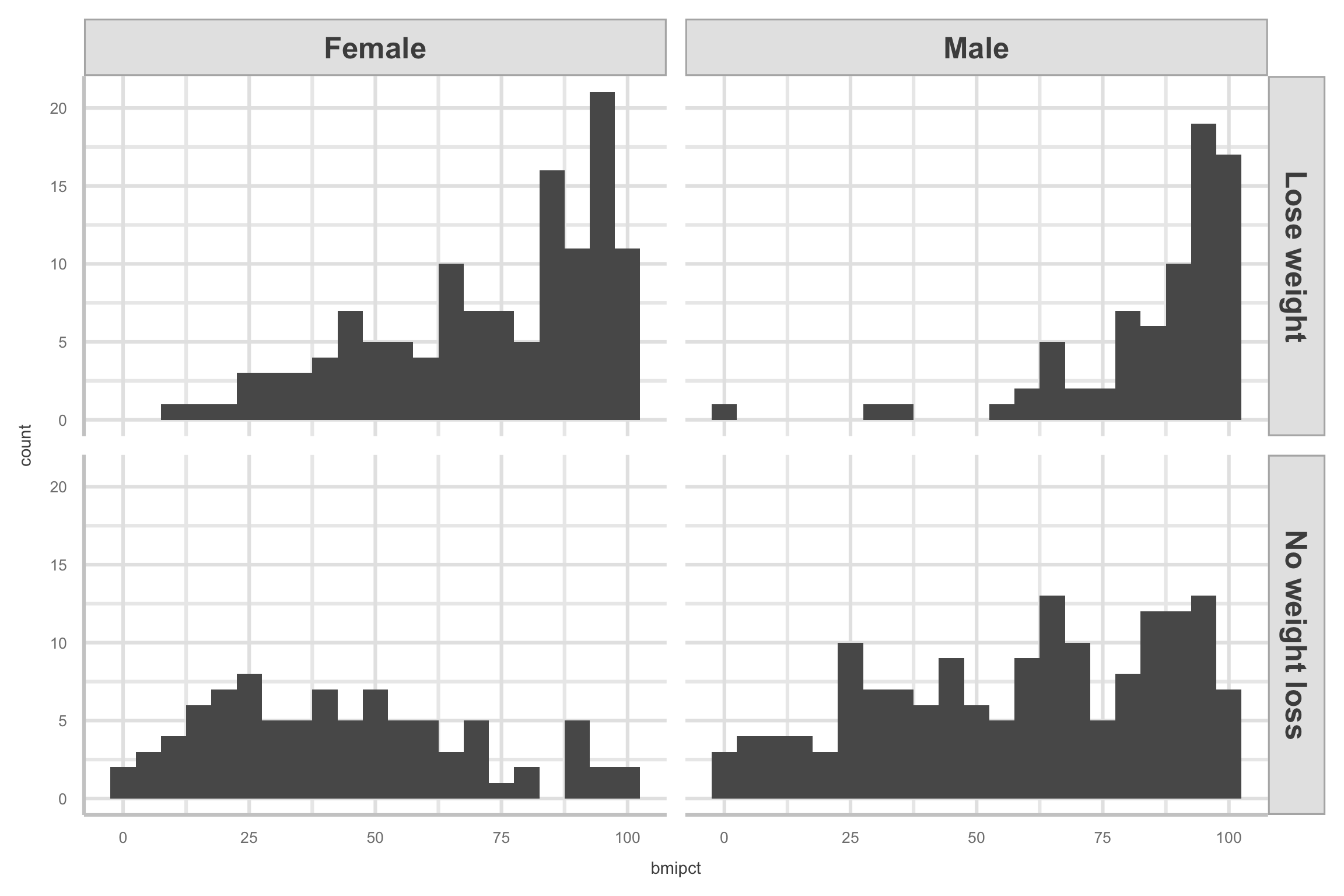
Logistic regression: Fitting model
- Let’s fit an intercept-only model
\[ \begin{align*} \text{lose.wt.01}_i & \sim \operatorname{Binomial}(n = 1, p_i)\\ \text{logit}(p_i) & = \beta_0 \end{align*} \]
Logistic Regression: Fitting Model
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -0.2120267 | 0.0953418 | -2.22386 | 0.0261579 |
Hypothesis test for \(\beta_j\)
Hypotheses:
\(H_0\): \(\beta_j\) = 0
\(H_a\): \(\beta_j \neq 0\)
Test Statistic: \[\text{Wald z} = \frac{\hat{\beta}_j - 0}{SE_{\hat{\beta}_j}}\]
CIs: \[\Large{\hat{\beta}_j \pm z^* SE_{\hat{\beta}_j}}\]
Interpreting \(b_0\)
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -0.2120267 | 0.0953418 | -2.22386 | 0.0261579 |
The intercept \(\beta_0\) is in the log-odds metric
- Log-odds of Y = 1 is -0.212
We can convert it to probability
- Thus, the overall probability of students wanting to lose weight was about .45 or 45%
Interpreting \(b_0\)
- Let’s check that with the empirical proportions
- We can actually do that transformation with the base R
plogis()function
Logistic Regression
- Add our binary predictor
female, which gives us the model
\[ \begin{align*} \text{lose.wt.01}_i & \sim \operatorname{Binomial}(n = 1, p_i) \\ \text{logit}(p_i) & = \beta_0 + \beta_1 \text{female}_i, \end{align*} \]
- Where
femaleis an 0/1 dummy variable.
Interpreting intercept: \(b_0\)
- \(b_0\):
Interpreting slope: \(b_1\)
If X is a categorical predictor
- Then \(b_1\) is the difference in the log-odds between group X and the baseline
- Going from men to women, there is a change in the the log odds of wanting to lose weight of 1.09
Coefficient effect size: Odd’s Ratio
We can take the
exp()of the log odds and get ORThe odds of Y for group X (females) are expected to be exp(\(b_1\)) times the odds of Y for the baseline group
Greater than 1: One unit increase, the event is more likely to occur
Less than 1: One unit increase, the event is less likely to occur
Equals 1: One unit increase, the likelihood of the event does not change
We can describe the OR in terms of percentage increase/decrease
Coefficient Effect Size: Odd’s Ratio
The odds of wanting to lose weight in females is 2.98 times the odds of wanting to lose weight in men
- The odds of females wanting to lose weight are 198% higher than the odds of males wanting to lose weight
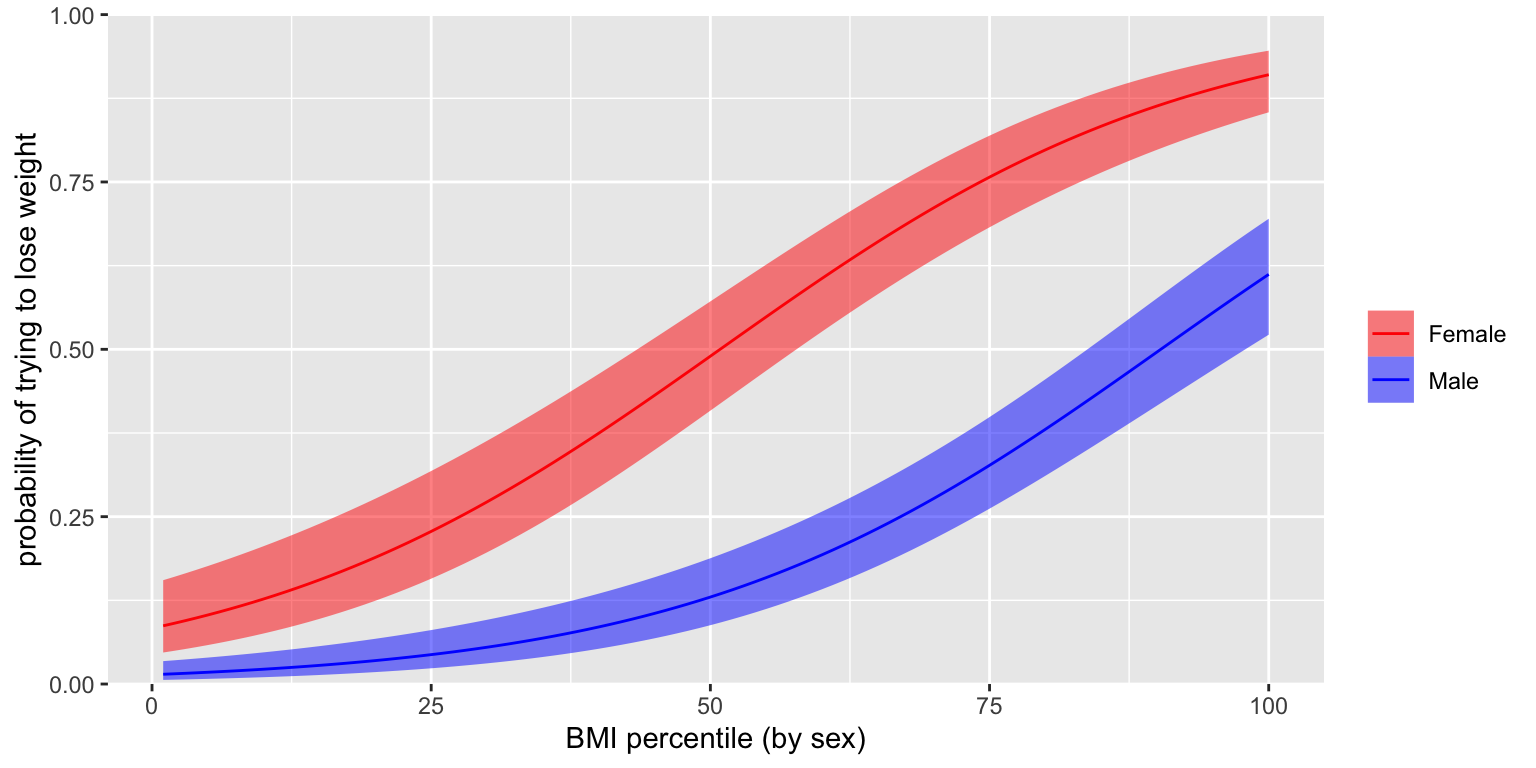
Predicted probabilities
- Now we can compute the probabilities for young men and women as follows
Emmeans
Can use
emmeansto get probabilities directly from model- Can also get odds ratios for comparisons!
female prob SE df asymp.LCL asymp.UCL 0 0.320 0.0307 Inf 0.263 0.383 1 0.584 0.0337 Inf 0.517 0.648 Confidence level used: 0.95 Intervals are back-transformed from the logit scalecontrast odds.ratio SE df null z.ratio p.value female1 / female0 2.98 0.589 Inf 1 5.520 <.0001 Tests are performed on the log odds ratio scale
Marginal effects
We can also calculate differences in probabilities
- Difference in probabilities is called risk difference
Marginal effects
Change in probability that the outcome occurs for a 1 unit change in the predictor
- Average marginal effect (AME)
- For categorical variables it is just probability difference between the group
#can do this with emmeans but requires more steps
library(marginaleffects) # get predicted probs
# get marginal effects
avg_comparisons(fit2, comparison = "difference") %>%
kable()| term | contrast | estimate | std.error | statistic | p.value | s.value | conf.low | conf.high |
|---|---|---|---|---|---|---|---|---|
| female | 1 - 0 | 0.2637658 | 0.0455817 | 5.786657 | 0 | 27.05335 | 0.1744273 | 0.3531044 |
Logistic regression continuous
Now we add our continuous predictor bmipct, which gives us the model:
\[ \begin{align*} \text{lose.wt.01}_i & \sim \operatorname{Binomial}(n = 1, p_i) \\ \text{logit}(p_i) & = \beta_0 + \beta_1 \text{bmipct}_i, \end{align*} \]
Logistic regression continuous
| term | estimate | std.error | statistic | p.value |
|---|---|---|---|---|
| (Intercept) | -2.5685641 | 0.3138083 | -8.185138 | 0 |
| bmipct | 0.0355475 | 0.0042828 | 8.300008 | 0 |
What is the OR for \(b_1\)? How do we interpret this?
Marginal effects
With continuous variables, obtaining the marginal effects is a bit trickier
Instantaneous rate of change in the predicted probability
- Change in the predicted probability for a very small change in X
#effect is computed for each observed value in the original dataset before being averaged
avg_slopes(fit3)%>%kable()| term | estimate | std.error | statistic | p.value | s.value | conf.low | conf.high |
|---|---|---|---|---|---|---|---|
| bmipct | 0.0071934 | 0.0005805 | 12.39111 | 0 | 114.7217 | 0.0060556 | 0.0083312 |
Tip
An increase in BMI results in 70% increase in likelihood of wanting to lose weight
Logistic Multiple Predictors
- The first will have both
femaleandbmipctas predictors:
\[ \begin{align*} \text{lose.wt.01}_i & \sim \operatorname{Binomial}(n = 1, p_i) \\ \text{logit}(p_i) & = \beta_0 + \beta_1 \text{female}_i + \beta_2 \text{bmipct}_i. \end{align*} \]
- The second model will add an interaction term between the two predictors:
\[ \begin{align*} \text{lose.wt.01}_i & \sim \operatorname{Binomial}(n = 1, p_i) \\ \text{logit}(p_i) & = \beta_0 + \beta_1 \text{female}_i + \beta_2 \text{bmipct}_i + \beta_3 \text{female}_i \cdot \text{bmipct}_i. \end{align*} \]
Fitting both models
Model Comparisons
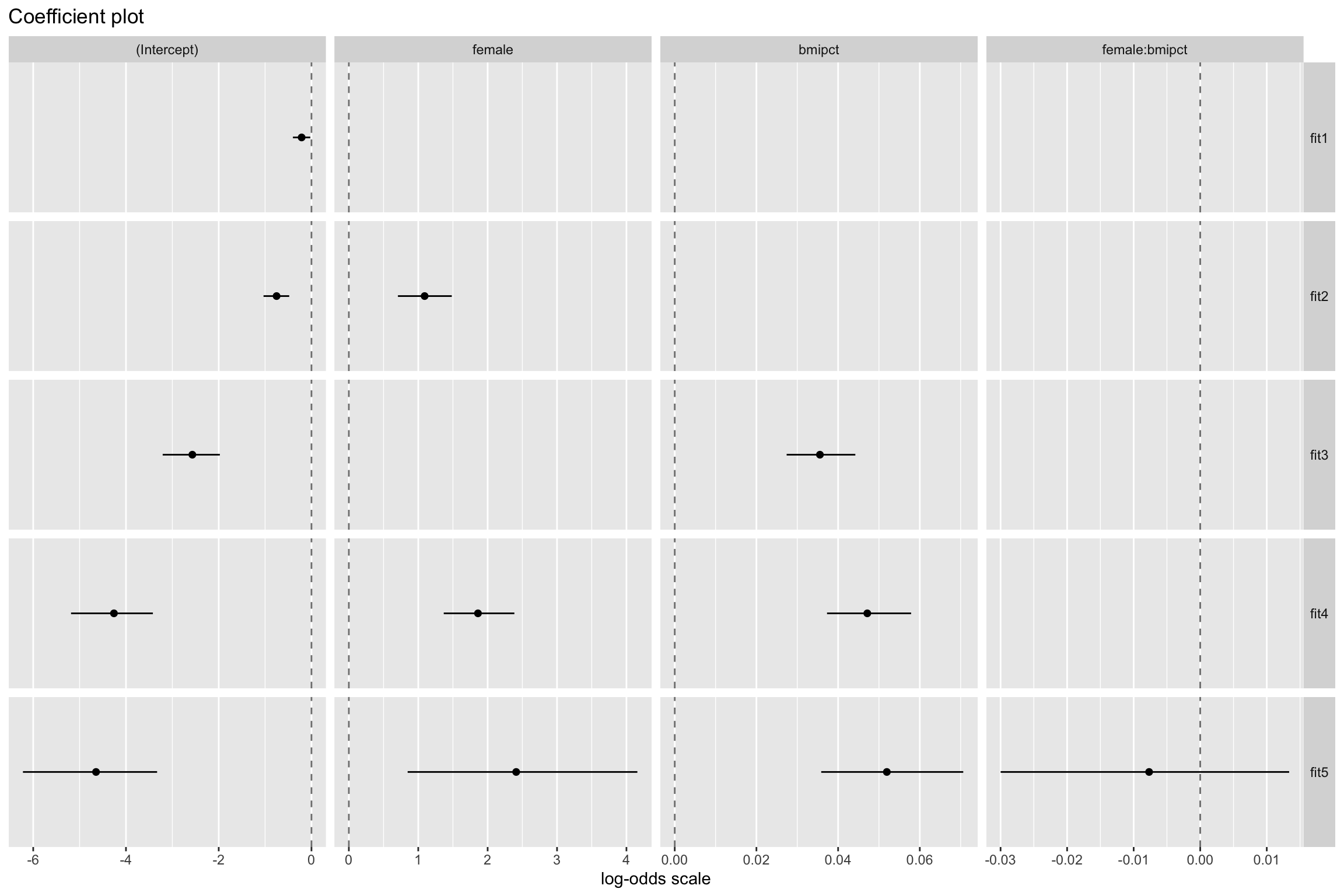
- The interaction term
female:bmipct(i.e., \(\beta_3\)) would not be statistically significant
Likelihood ratio test
- Let’s compare the models by performing likelihood ratio test
| Name | Model | df | df_diff | Chi2 | p | |
|---|---|---|---|---|---|---|
| fit4 | fit4 | glm | 3 | NA | NA | NA |
| fit5 | fit5 | glm | 4 | 1 | 0.4976291 | 0.4805437 |
- When you compare model
fit4with the interaction modelfit5, it appears that the interaction term does not add anything
Model Fit
- McFadden Psuedo \(R^2\)
\[R^2 = \frac{SSM}{SST} = \frac{SST-SSE}{SST}=1-\frac{SSE}{SST}\] \[R_{MF}^2 = 1-\frac{D_{\rm residual}}{D_{\rm null}}\]
# R2 for Generalized Linear Regression
R2: 0.243
adj. R2: 0.240- Not “explained variance” but ratio indicating how “good” the model fit Model Fit
Model fit
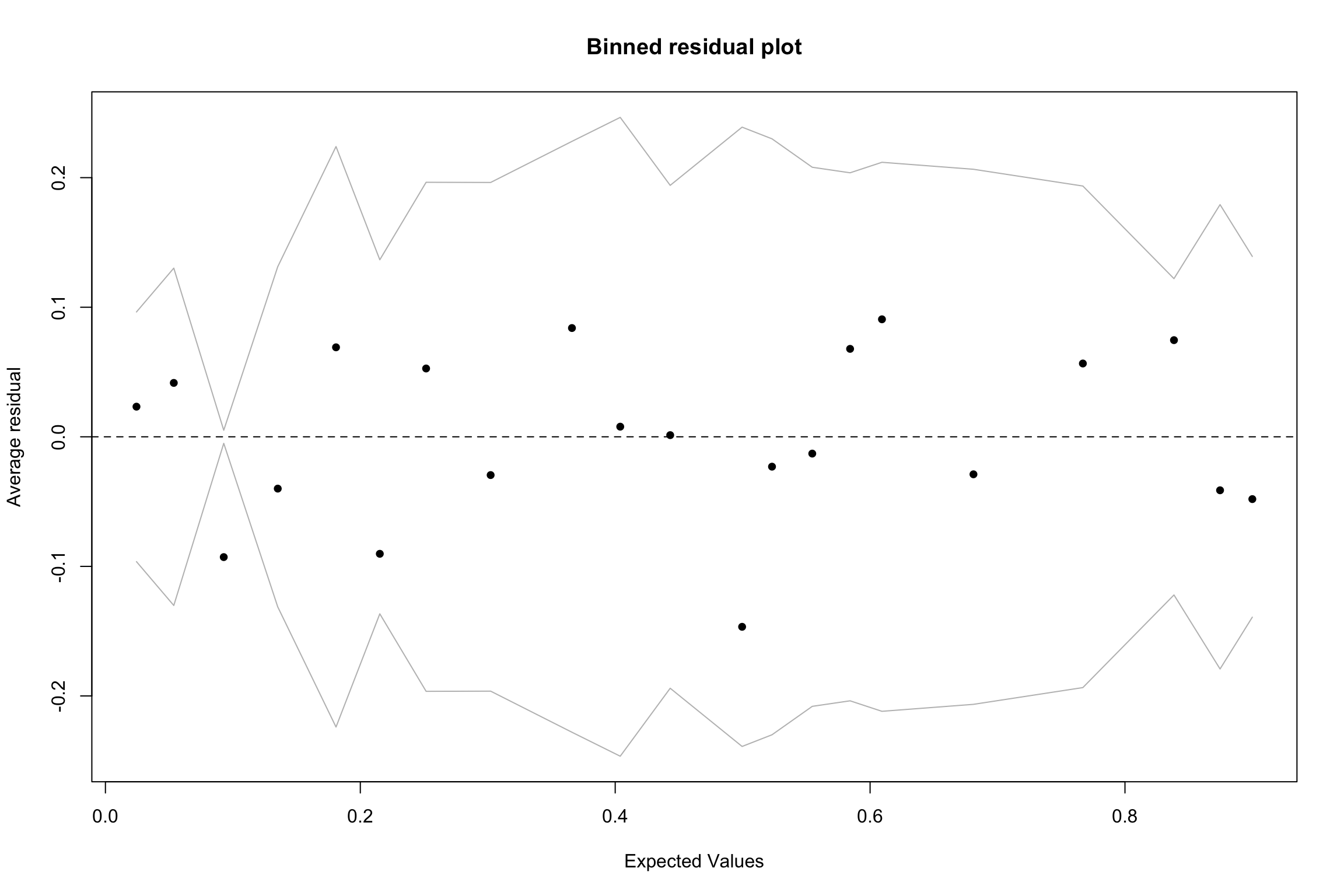
- Plotting normal residuals are not really informative/useful
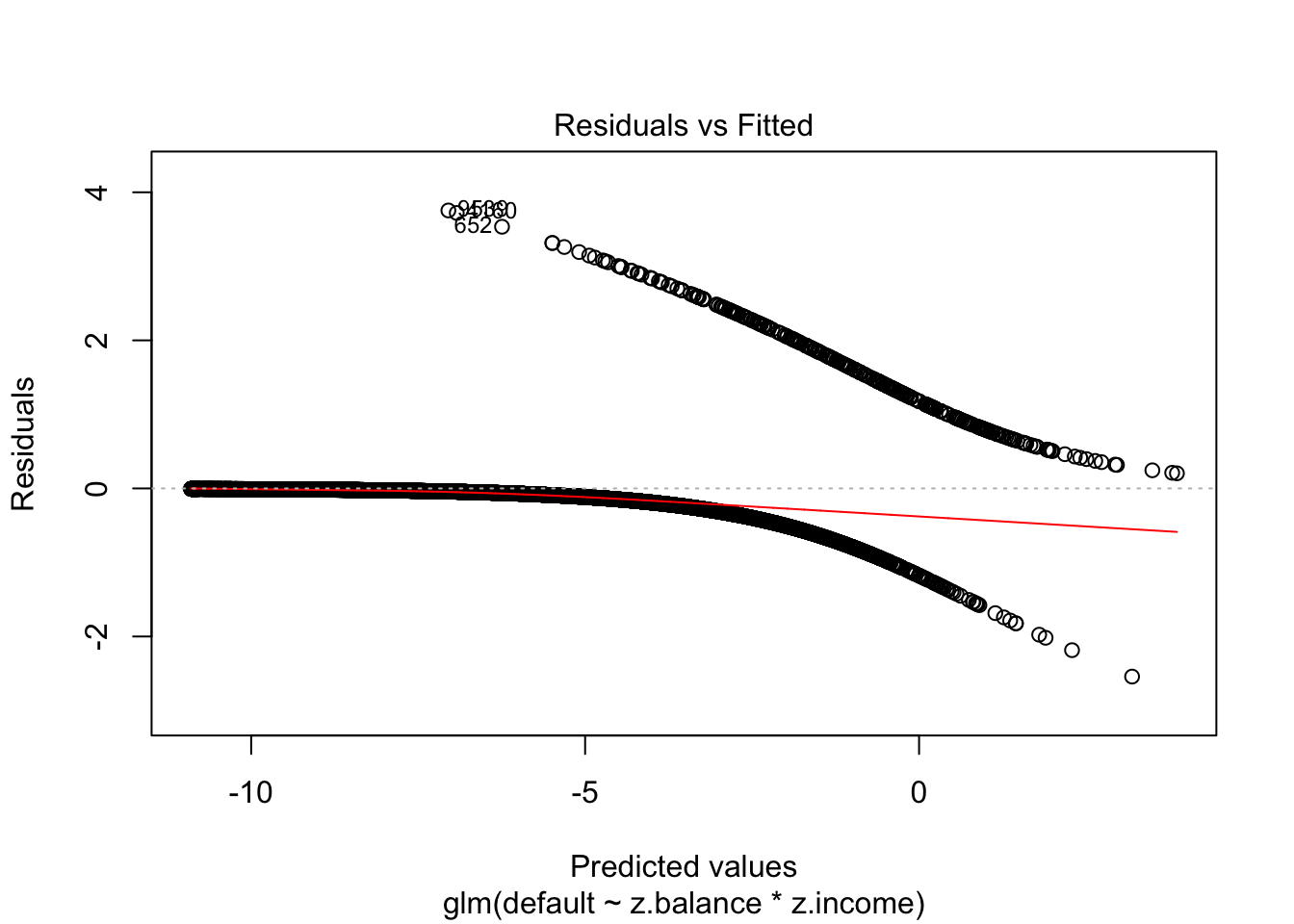
Binned residuals
Binned residuals
- Calculate raw residuals
- Order observations (probabilities of DV or predictor variable)
- Create g bins of approximate equal size \(\sqrt(n)\)
- Calculate average residuals
- Plot average residuals vs. average predicted probability (or predictor value)
Binned residuals
Look for patterns
- Nonlinear trend may be indication that squared term or log transformation is required
If bins have average residuals with large magnitude
Look at averages of other predictors across bins
Interaction may be required
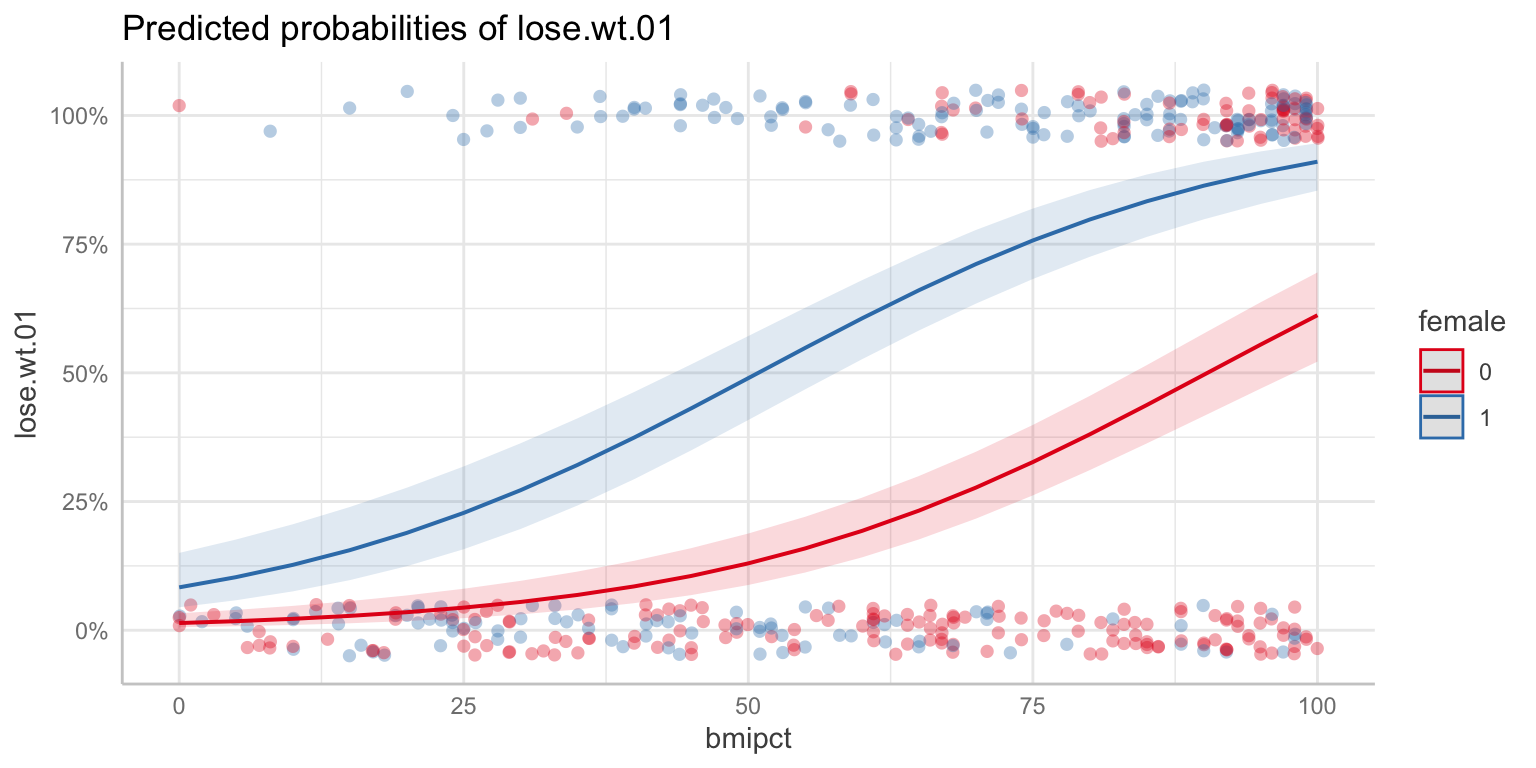
Visualizing
Assumptions
Logistic regression assumptions
Outcome is dichotomous
Observations are independent
Linearity (in the log odds)
No normality assumptionsHomogeneity of variance- Variance varies with \(p\)
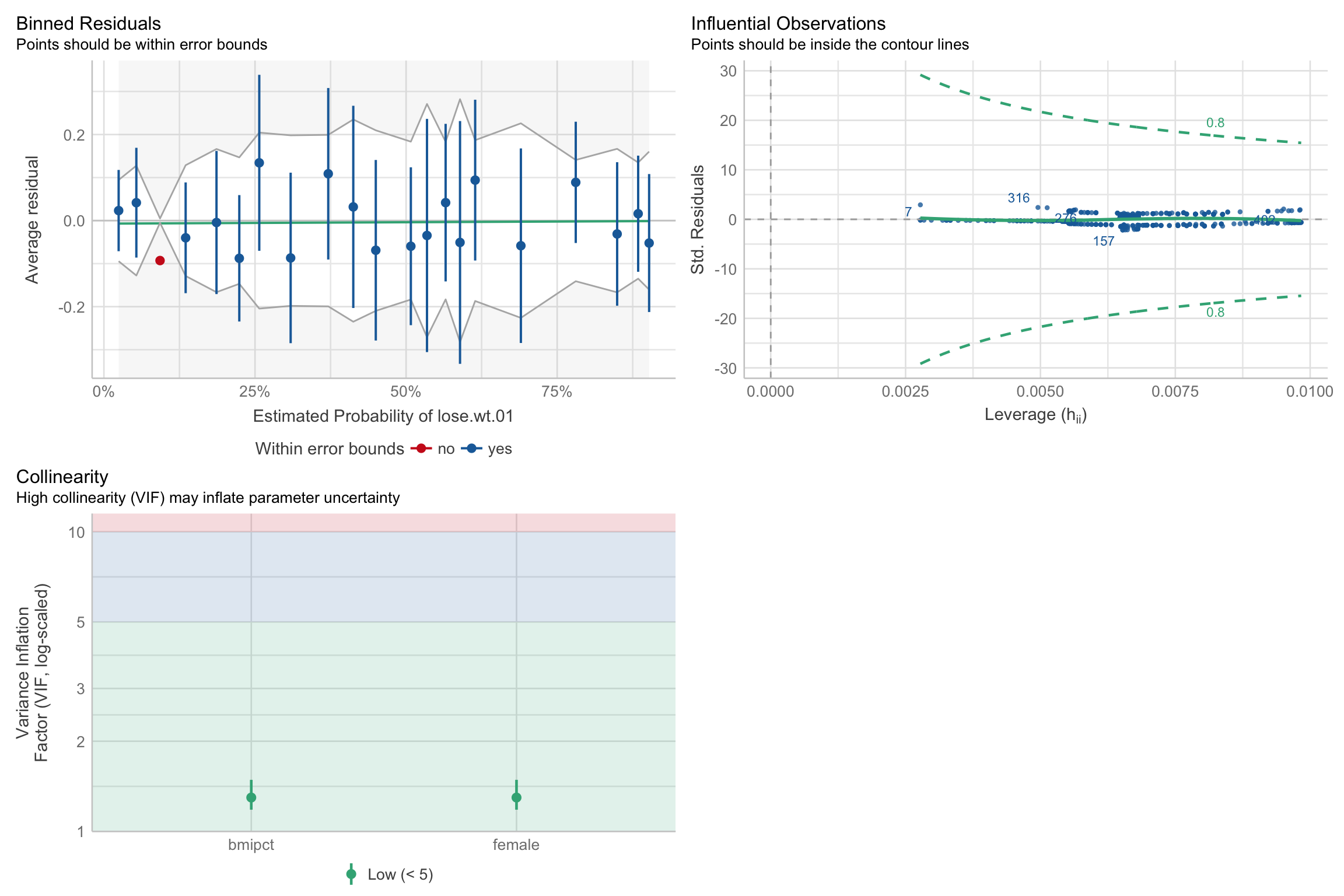
Logistic regression assumptions
Can plot with
easystats:)Note
- Should check binned residuals of the predictor variables as well
Table
Reporting
Tip
We fitted a logistic model with a logit link (estimated using ML) with sex and bmipct predicting desire to lose weight (formula: lose.wt.01 ~ 1 + female+ bmipct). The model’s explanatory power is substantial (Tjur’s \(R^2\) =0.30). The probability of females wanting to lose weight was 33% higher, b= 1.86, 95% CI [1.37, 2.38], p < .001; OR = 2.98, 95% CI [3.93, 10.9]. An increase in BMI was associated with an increased probability of wanting to lose weight, b = 0.05, 95% CI [0.04, 0.06], p < .001, OR = 1.05, 95% CI [1.04, 1.06]. Looking at the average marginal effect, 1 unit increases in BMI was associated with a 80% increase in desire to lose weight.
PSY 504: Advanced Statistics