Introduction to Structural Equation Modeling in R
Princeton University
2024-04-14
Today
What is structural equation modeling (SEM)?
Path models
Important terminology
How to do it in R
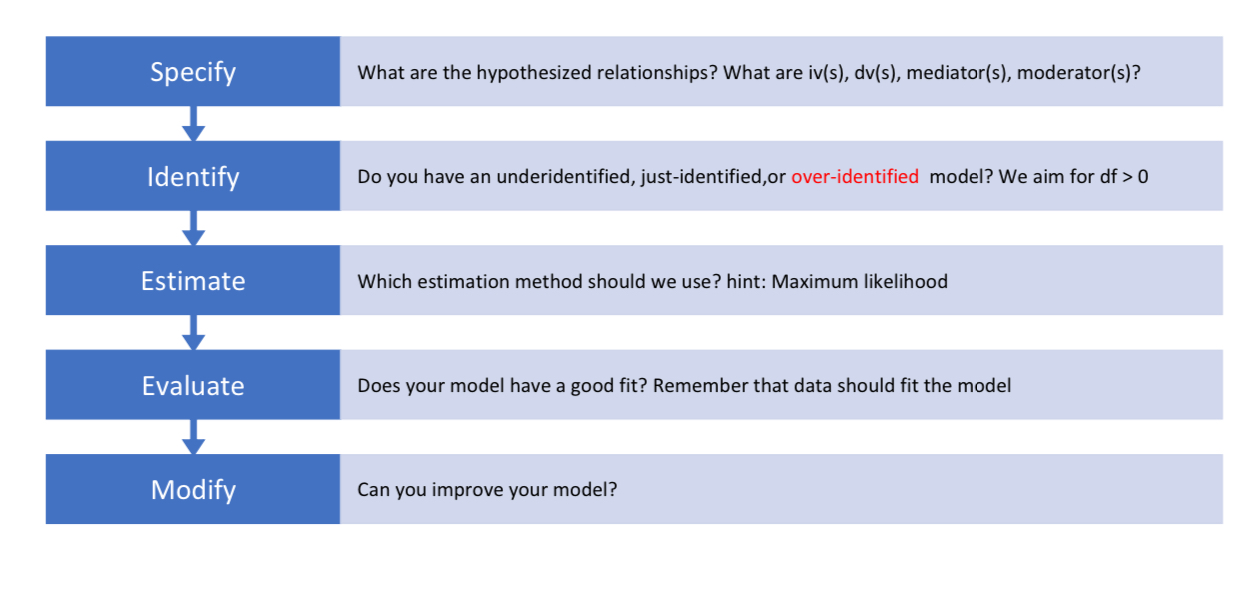
Specification
Identification
Estimation
Model fit & indices
Modification indices
Reporting
Packages
Structural equation modeling
A broad range of of techniques and frameworks
Not a single technique
Integration of:
Path analysis
Confirmatory factor analysis
Used to test and quantify theories
Model variables that exist (manifest) and those that don’t technically exist (latent factors)
Structural equation modeling

You already know how to do it!
It is regression on steroids
Model many relationships at once, rather than run single regressions
Path analysis
- A method for testing any possible relationship between measured variables
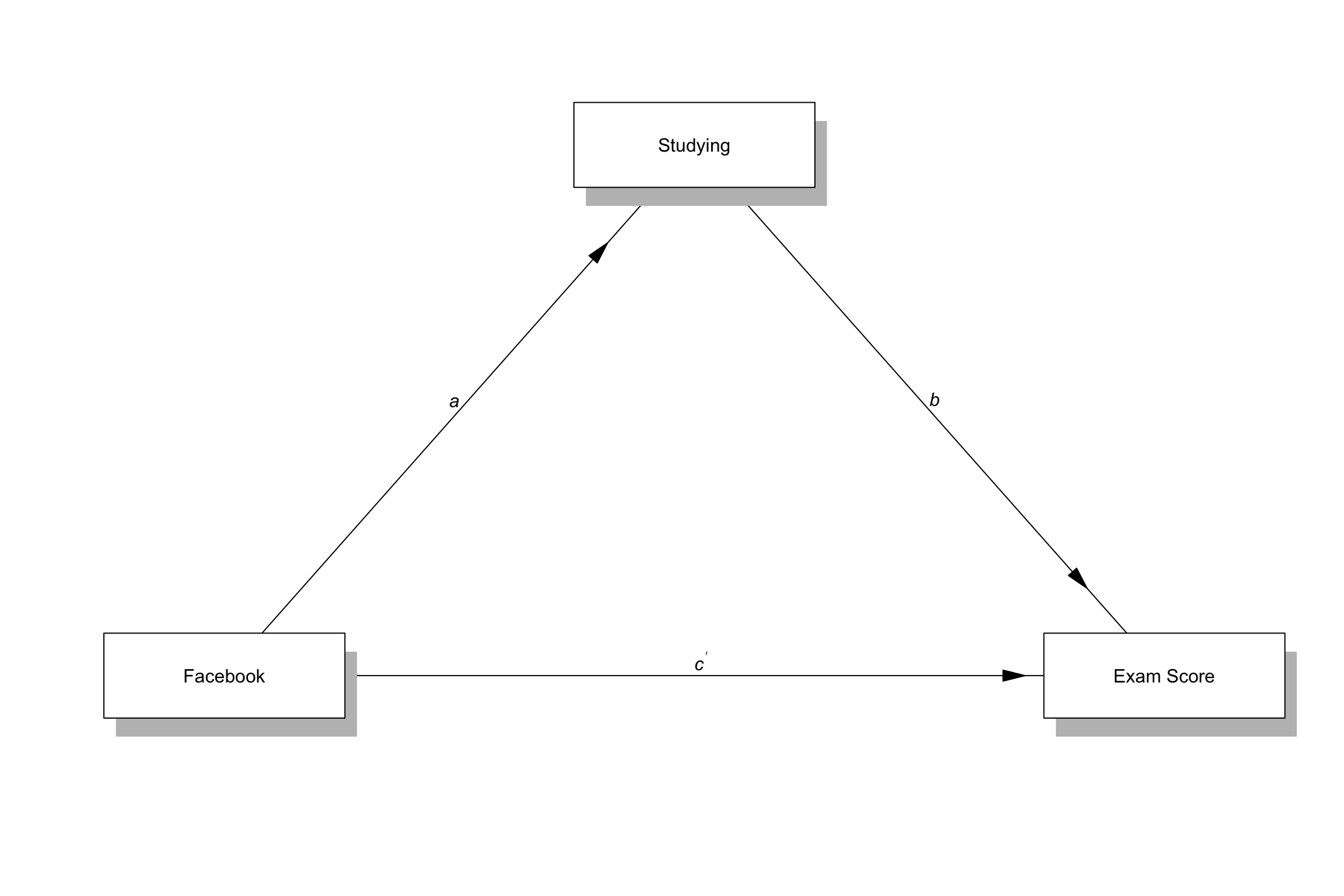
Terminology
Exogenous vs. endogenous variables
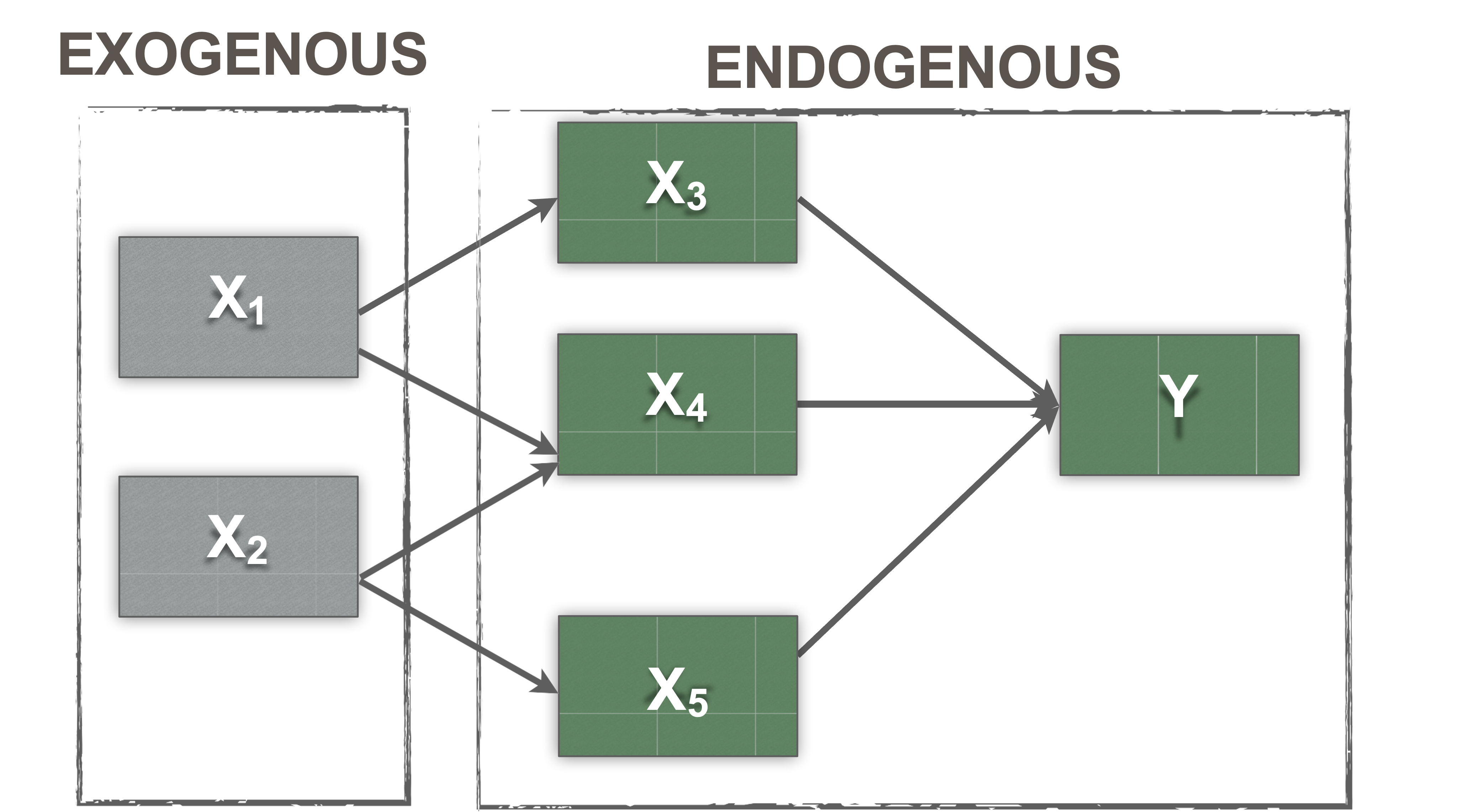
- Exogenous
- These are synonymous with independent variables
- You can find these in a model where the arrow is leaving the variable
- They are thought to be the cause of something
- Have variance
- Covary with other exogenous variables (default)
Exogenous vs. endogenous variables

- Endogenous
- These are synonymous with dependent variables
- They are caused by the exogenous variables
- In a model diagram, the arrow will be coming into the variable
- Have error terms (disturbances)
Endogenous vs. Exogenous
TL;DR
Exogenous: no arrows pointed at it
Endogenous: arrows pointed at it
Manifest variables vs. latent variables

Manifest or observed variables
Represented by squares ❏
Measured from participants, business data, or other sources
While most measured variables are continuous, you can use categorical and ordered measures as well
Manifest variables vs. latent variables
Latent variables
Represented by circles ◯
Abstract phenomena you are trying to model
Are not represented by a number in the dataset
Linked to the measured variables
Represented indirectly by those variables
Remember

Y~X + ResidualHere that is
Endogenous ~ Exogenous + disturbance
Variances, covariances, and disturbances
- Variance
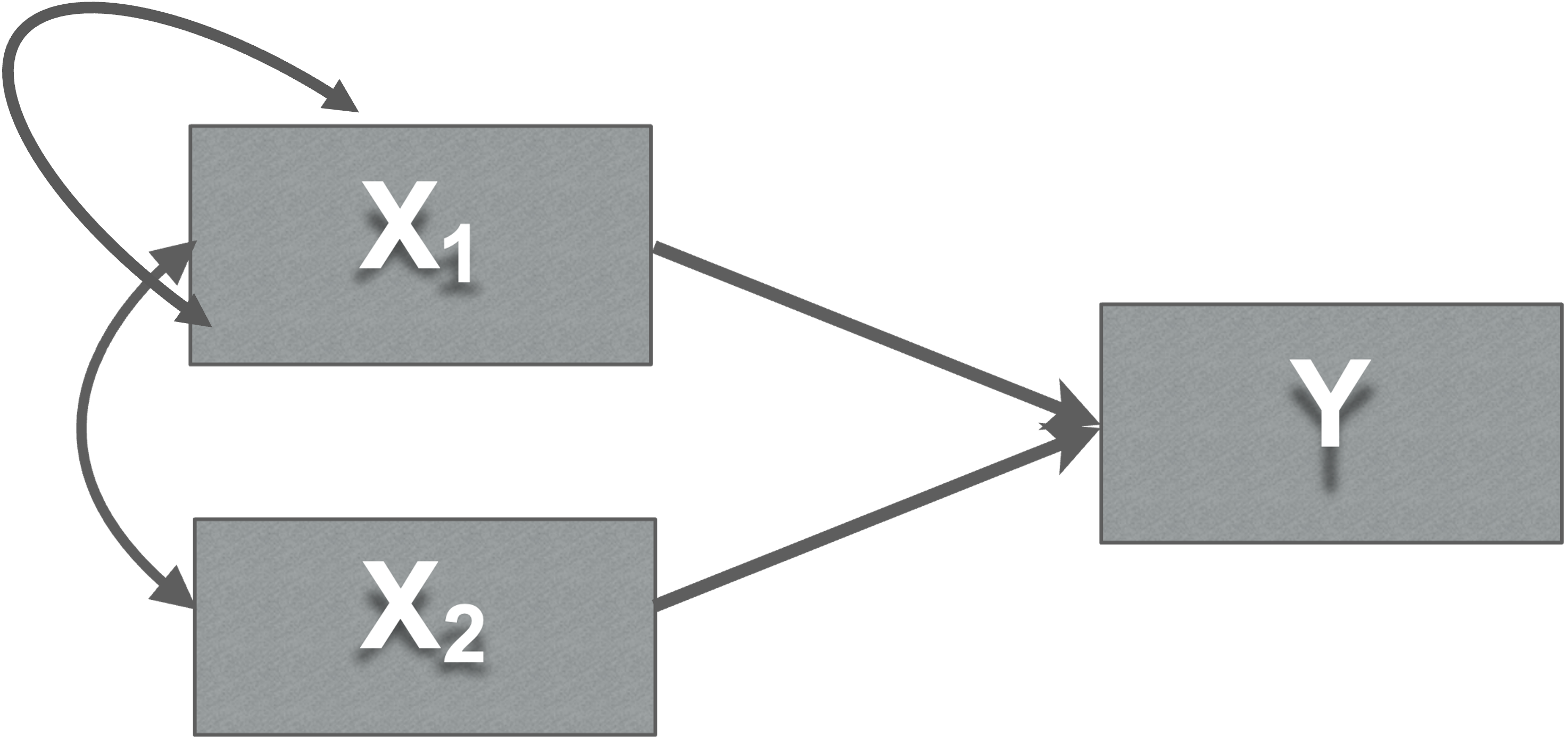
- Double headed arrows
- To itself
Variances, covariances, and disturbances
- Covariance paths

- Double headed arrows (Covariance paths)
- Exogenous variables may be correlated with each other, but not always…
Covariance meaning
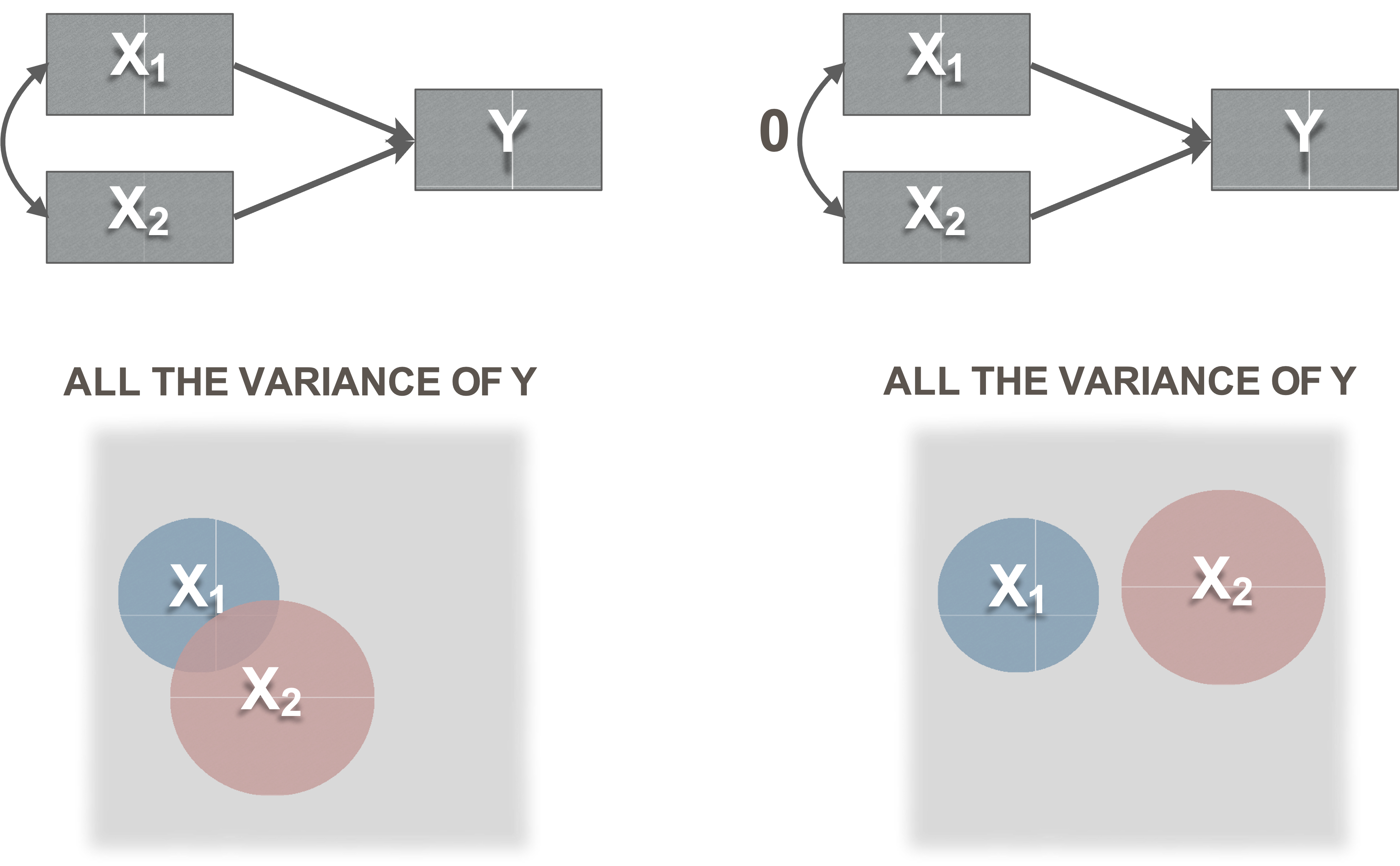
Variances, covariances, and disturbances
- Disturbances
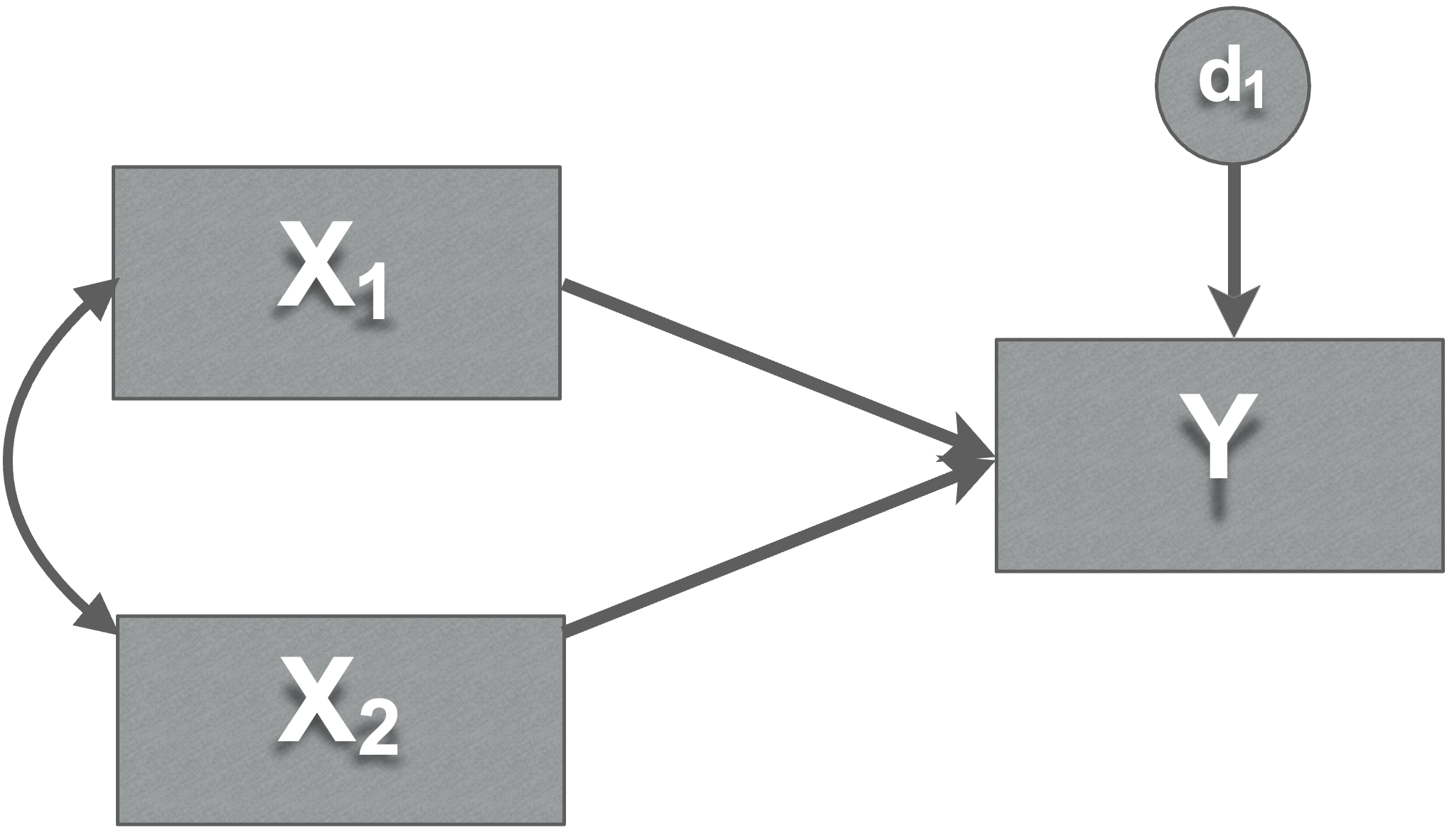
Represent the influence of factors not included in model
error in your prediction of each endogenous variable
Every endogenous variable has a disturbance
SEM models
Straight arrows are “causal” or directional
- Non-standardized solution -> these are your b or slope values
- Standardized solution -> these are your beta values
Curved arrows are non-directional
- Non-standardized -> covariance
- Standardized -> correlation
Cheat sheet
- Also here: https://davidakenny.net/cm/basics.htm
Why is it just regression?
Each endogenous variable is regressed on all exogenous variables that are connected in the chain that leads directly to it
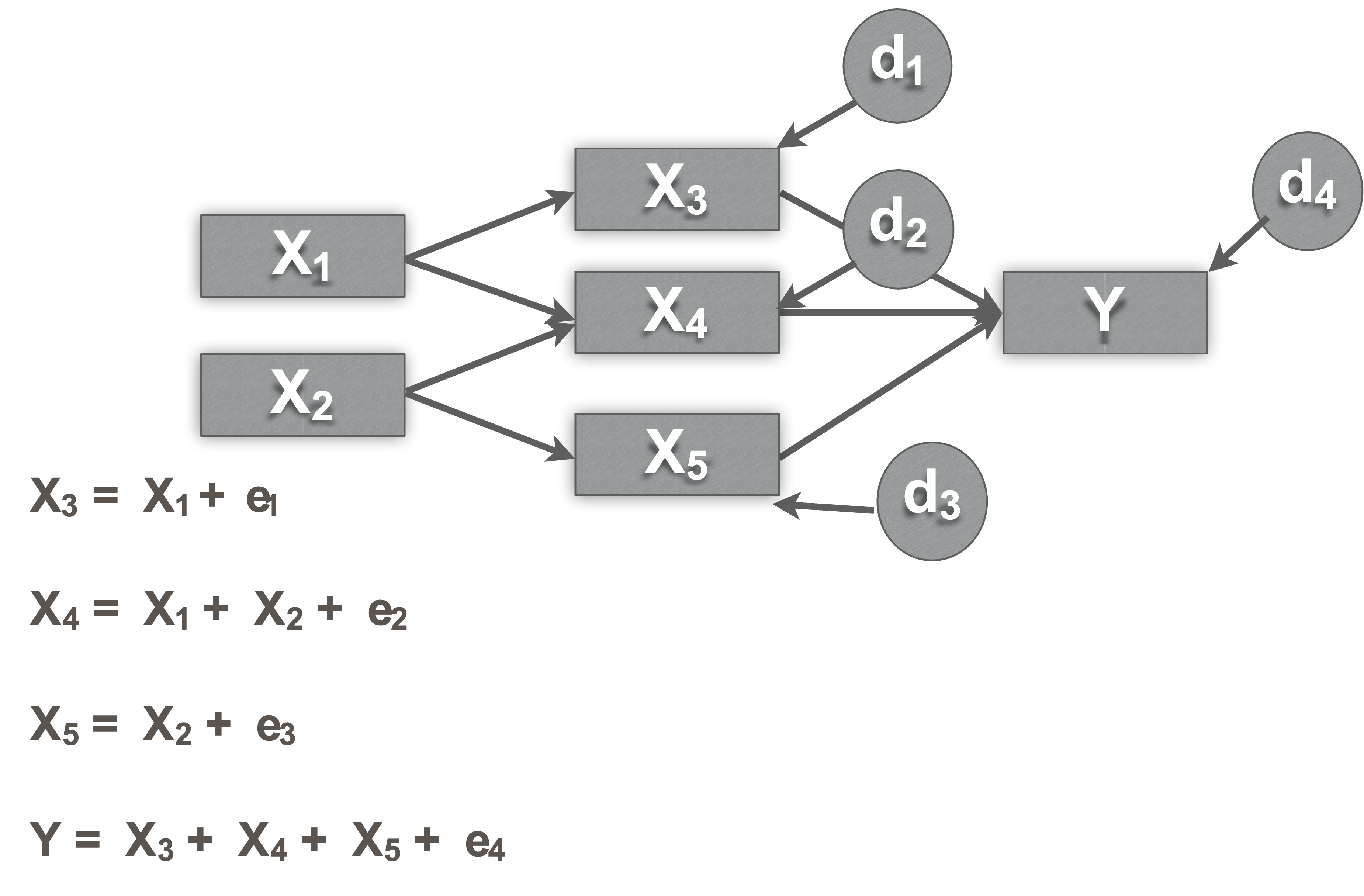
Identification
Model identification
You cannot test all models
- Unique solution
If not identified, cannot analyze model
How is this determined?
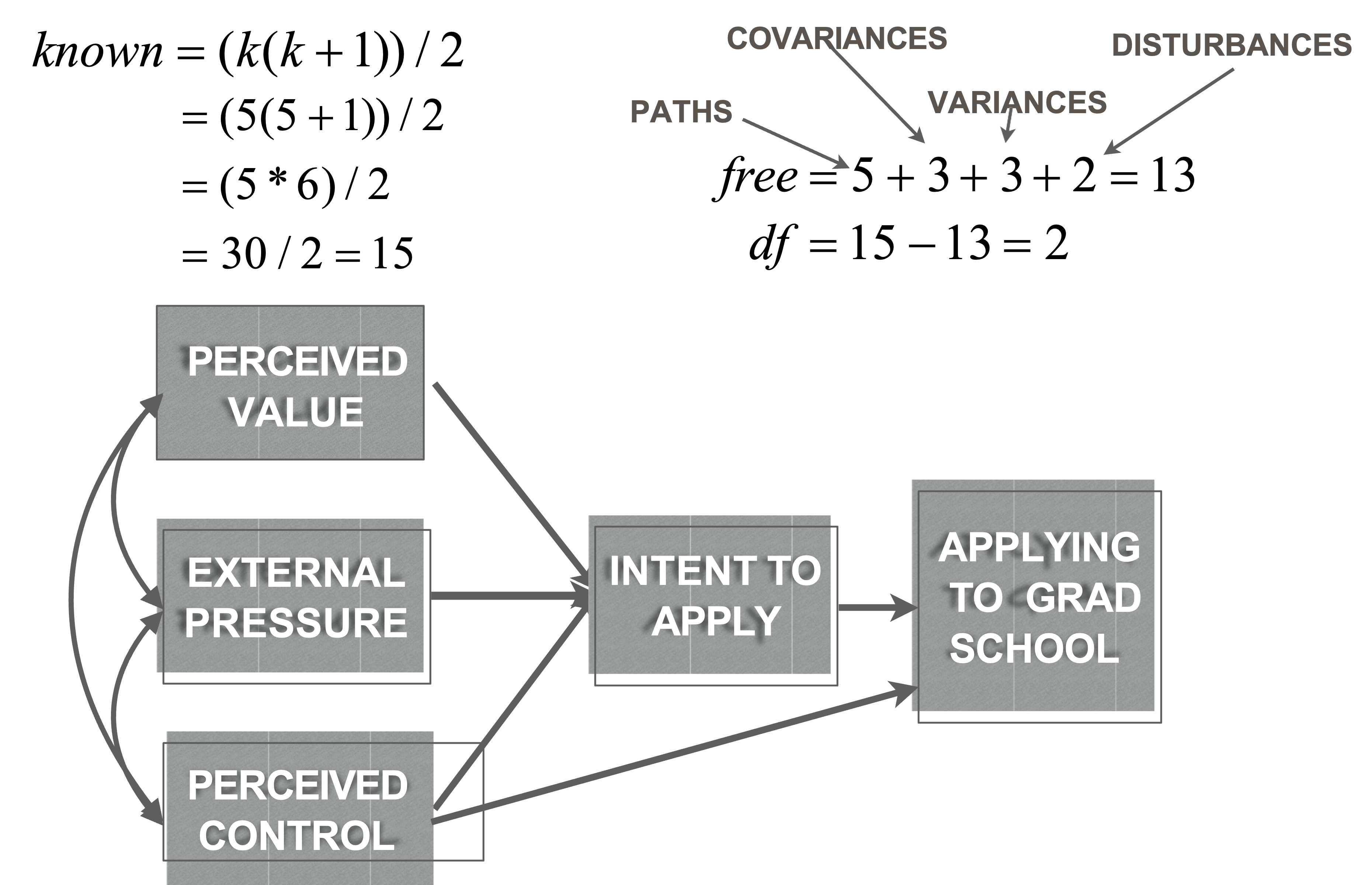
Minimum condition of identification
There must be at least as many known values in the model as there are free parameters
Free parameters:
- All regression paths, all covariances, all variances, and all disturbances in the model
Knowns:
\[\frac{(K (K+1))}{2}\]- where k is number of measured variables
Model identification
We can tell model is identified by calculating model DFs
- Additional pathways you can estimate
Model DF = (known values) - (free parameters)
Model identification
If model DF >=1 you can analyze model
- Over-identified
DF = 0 (saturated)
You can still analyze the model
But:
Fits data perfectly
No fit indices
Multiple regressions are just-identified model
DF < 0
Under-identified
Cant analyze our model
Identification
Just-identified - mathematical analogy
10=2x+y
2 = x-y
2 knows and 2 unknowns
One set of values that can solve the equation
Can’t test other models
Under-identified - mathematical analogy
10 = x+ y
One known value and 2 unknowns (x,y)
There are infinite number of solutions
No way to derive a solution
Identification
Over-identified - mathematical analogy
10=2x+y
2 = x-y
5 = x + 2y
- Several possible approximate solutions to solve all 3 equations
- Several unique but imperfect solutions means multiple models can be tested or compared
- You can’t evaluate fit without alternatives

Estimate DFs
- Let’s play a game

03:00
SEM steps
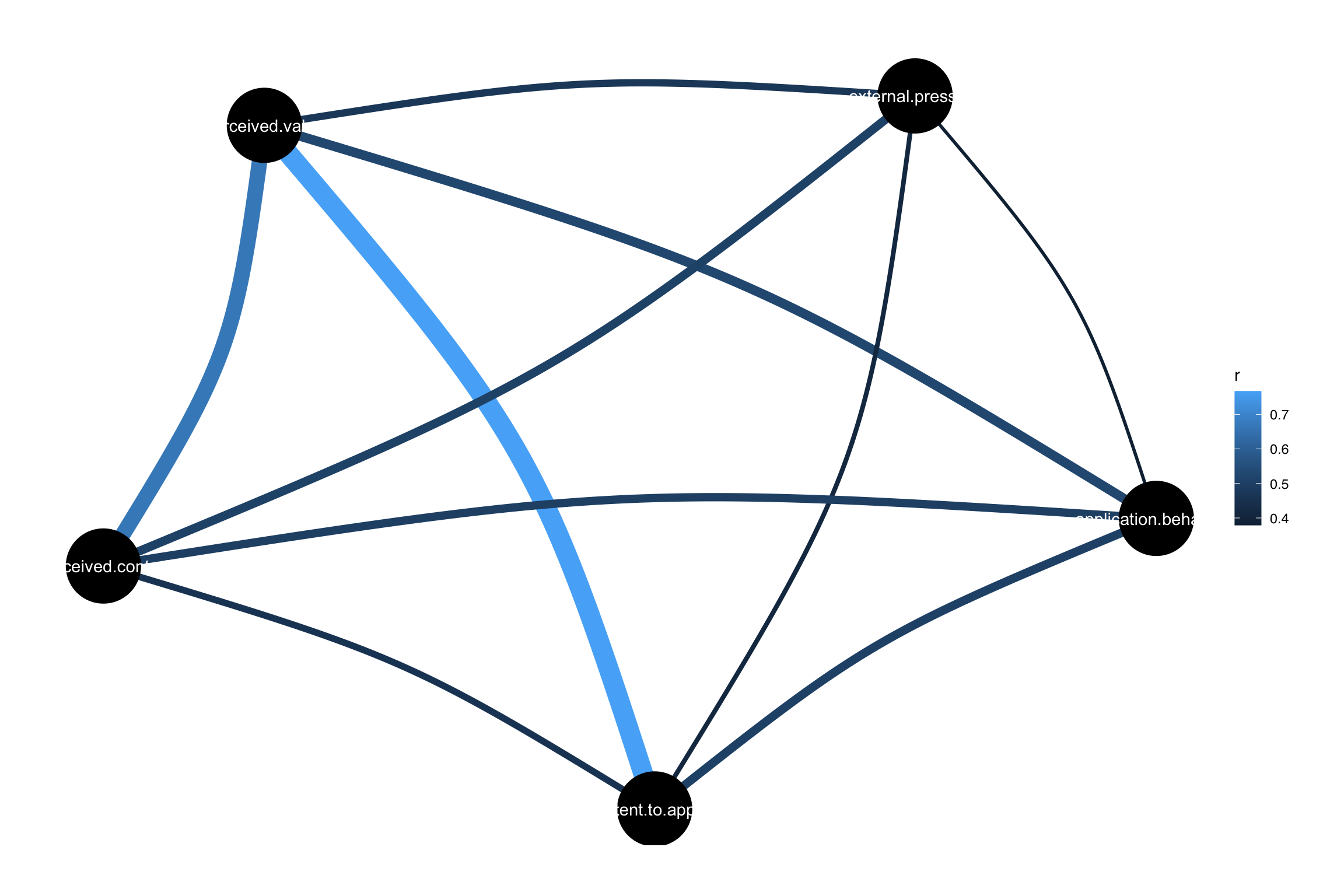
Example data (Ingram et al., 2000)
What makes someone apply to graduate school?
Endogenous
- Intent to apply (intent.to.apply)
- Apply (application.behaviour)
Exogenous:
- Perceived value (perceived.value)
- External pressure (external.pressure)
- Perceived control over admission (perceived.control)
Model specification
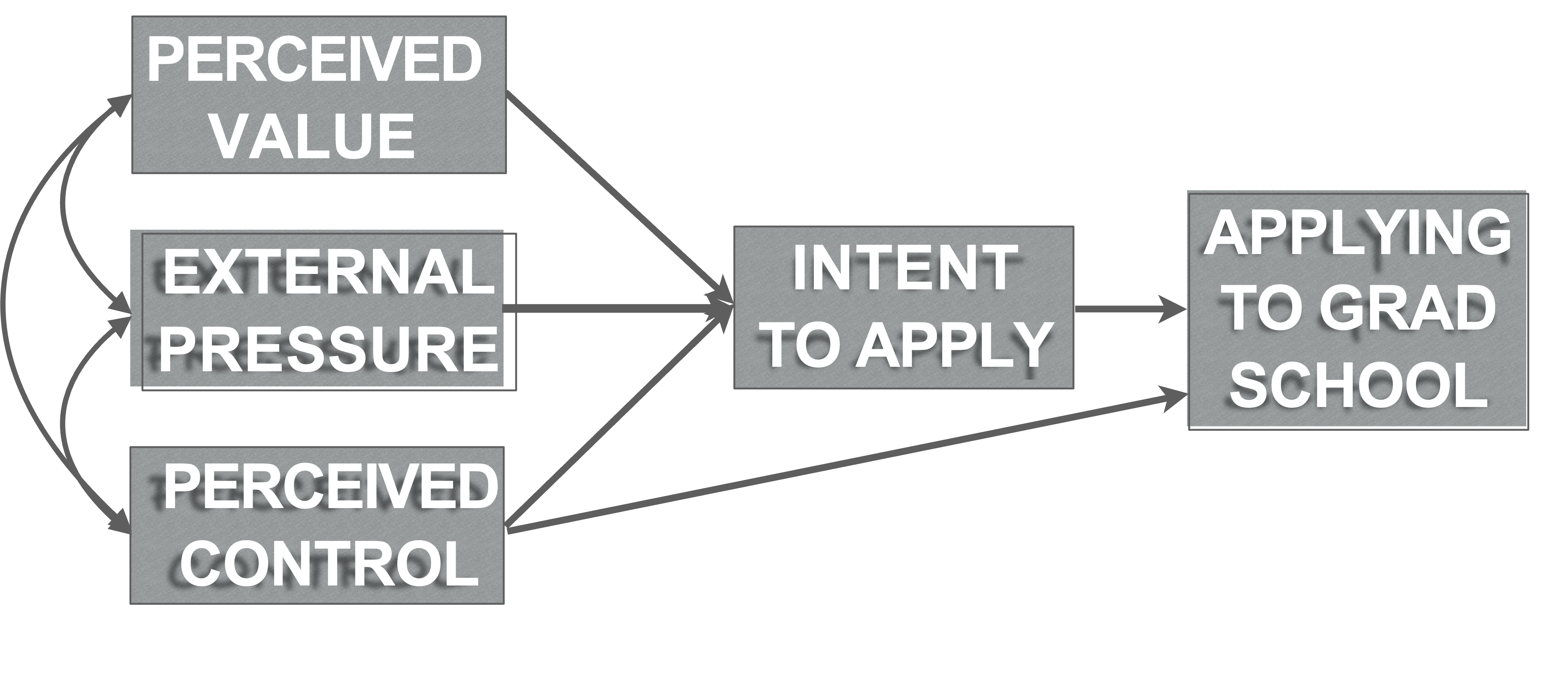
- Theory of planned behavior
Data screening
Identification
Run SEM in R
Declare equations for every endogenous variable in your model
- `~` indicates a regression
Declare indirect and covariances
:=declare a variable*name of variable~~indicates a covariance/correlation=~latent factor
Run SEM in R
grad_model = '
intent.to.apply~a*perceived.value+b*external.pressure+c*perceived.control
application.behaviour~d*intent.to.apply+ e*perceived.control
#indirect
value.through.intent:=a*d
#indirect
pressure.through.intent:=b*d
#indirect
control.through.intent:=c*d
perceived.control ~~ perceived.value # These are covariance paths
perceived.control ~~ external.pressure # These are covariance paths
external.pressure ~~ perceived.value # These are covariance paths
'
fit <- sem(grad_model, se="bootstrap", bootstrap=5000, data=grad)Model fit
SEM compares the observed covariance matrix to predicted or model-based covariance matrix
We want to know if our theoretical model fits the data
Good fit means we’ve captured the bulk of relations between variables in our model and there is not much covariance left over
- We didn’t miss anything
This means we compare the relationships we’ve modeled to all possible relationships
Model fit
| Fit measure | Name | Cutoff |
|---|---|---|
| \(\chi^2\)* | Chi-square | p > .05 |
| (A)GFI | Adjusted Goodness of Fit | AGFI ≥ .90 |
| TLI* | Tucker Lewis Index | TLI ≥ .95 |
| CFI* | Comparative Fit Index | CFI ≥ .90 |
| RMSEA* | Root Mean Square Error of Approximation | RMSEA < .08 |
| (S)RMR* | Standardized Root Mean Square Residual | SRMR < .08 |
| AVE (in CFAs) | Average Value Explained | AVE > .5 |
- Common to use \(\chi^2\), RMSEA, SRMR, CFI
Note
Hu, L.-t., & Bentler, P. M. (1999). Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Structural Equation Modeling, 6(1), 1–55. https://doi.org/10.1080/10705519909540118
Model fit
Absolute fit (measures how well data fits specified model)
\(\chi^2\) (sensitive to sample size)
SRMR
- Standardized difference between the sample covariance matrix and hypothesized covariance matrix
Badness of fit
RMSEA
- Measures how much worse the data fits the model from the just identified model
Relative goodness of fit
CFI or TLI
- Measures how much better the model fits than the null model (all paths=0)
Run SEM
fit <- sem(grad_model, se="bootstrap", bootstrap=5000, data=grad)
summary(fit, ci=TRUE,
standardize=TRUE, # get standardized
fit.measures=TRUE) # get fit indiceslavaan 0.6.17 ended normally after 60 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 13
Number of observations 60
Model Test User Model:
Test statistic 0.862
Degrees of freedom 2
P-value (Chi-square) 0.650
Model Test Baseline Model:
Test statistic 136.416
Degrees of freedom 10
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 1.000
Tucker-Lewis Index (TLI) 1.045
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -993.733
Loglikelihood unrestricted model (H1) -993.303
Akaike (AIC) 2013.467
Bayesian (BIC) 2040.693
Sample-size adjusted Bayesian (SABIC) 1999.805
Root Mean Square Error of Approximation:
RMSEA 0.000
90 Percent confidence interval - lower 0.000
90 Percent confidence interval - upper 0.200
P-value H_0: RMSEA <= 0.050 0.690
P-value H_0: RMSEA >= 0.080 0.257
Standardized Root Mean Square Residual:
SRMR 0.019
Parameter Estimates:
Standard errors Bootstrap
Number of requested bootstrap draws 5000
Number of successful bootstrap draws 4998
Regressions:
Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
intent.to.apply ~
percevd.vl (a) 0.444 0.058 7.714 0.000 0.323 0.551
extrnl.prs (b) 0.029 0.035 0.850 0.396 -0.035 0.101
prcvd.cntr (c) -0.064 0.059 -1.075 0.282 -0.191 0.042
application.behaviour ~
intnt.t.pp (d) 1.520 0.535 2.840 0.005 0.487 2.593
prcvd.cntr (e) 0.734 0.303 2.422 0.015 0.249 1.455
Std.lv Std.all
0.444 0.807
0.029 0.095
-0.064 -0.126
1.520 0.350
0.734 0.336
Covariances:
Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
perceived.value ~~
perceivd.cntrl 34.696 10.891 3.186 0.001 16.025 58.051
external.pressure ~~
perceivd.cntrl 46.660 13.080 3.567 0.000 22.627 73.386
perceived.value ~~
external.prssr 39.758 11.894 3.343 0.001 16.815 63.559
Std.lv Std.all
34.696 0.665
46.660 0.505
39.758 0.472
Variances:
Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
.intent.to.pply 5.780 0.987 5.855 0.000 3.537 7.384
.applicatn.bhvr 179.514 27.923 6.429 0.000 118.564 228.012
perceived.valu 47.616 9.468 5.029 0.000 30.203 67.277
external.prssr 149.236 22.301 6.692 0.000 105.792 193.454
perceivd.cntrl 57.154 14.014 4.078 0.000 31.636 85.717
Std.lv Std.all
5.780 0.400
179.514 0.657
47.616 1.000
149.236 1.000
57.154 1.000
Defined Parameters:
Estimate Std.Err z-value P(>|z|) ci.lower ci.upper
val.thrgh.ntnt 0.675 0.252 2.683 0.007 0.200 1.199
prssr.thrgh.nt 0.045 0.058 0.768 0.442 -0.055 0.180
cntrl.thrgh.nt -0.097 0.111 -0.870 0.384 -0.379 0.056
Std.lv Std.all
0.675 0.282
0.045 0.033
-0.097 -0.044What to report?
easystatshas you covered!
Name Value Threshold Interpretation
1 GFI 0.99432949 0.95 satisfactory
2 AGFI 0.95747118 0.90 satisfactory
3 NFI 0.99368415 0.90 satisfactory
4 NNFI 1.04502659 0.90 satisfactory
5 CFI 1.00000000 0.90 satisfactory
6 RMSEA 0.00000000 0.05 satisfactory
7 SRMR 0.01902056 0.08 satisfactory
8 RFI 0.96842073 0.90 satisfactory
9 PNFI 0.19873683 0.50 poor
10 IFI 1.00846935 0.90 satisfactoryReporting Results
The model is not significantly different from a baseline model (Chi2(2) = 0.86,
p = 0.650). The GFI (.99 > .95) suggest a satisfactory fit. The PNFI (.20 <
.50) suggests a poor fit., The model is not significantly different from a
baseline model (Chi2(2) = 0.86, p = 0.650). The AGFI (.96 > .90) suggest a
satisfactory fit. The PNFI (.20 < .50) suggests a poor fit., The model is not
significantly different from a baseline model (Chi2(2) = 0.86, p = 0.650). The
NFI (.99 > .90) suggest a satisfactory fit. The PNFI (.20 < .50) suggests a
poor fit., The model is not significantly different from a baseline model
(Chi2(2) = 0.86, p = 0.650). The NNFI (.05 > .90) suggest a satisfactory fit.
The PNFI (.20 < .50) suggests a poor fit., The model is not significantly
different from a baseline model (Chi2(2) = 0.86, p = 0.650). The CFI (.00 >
.90) suggest a satisfactory fit. The PNFI (.20 < .50) suggests a poor fit., The
model is not significantly different from a baseline model (Chi2(2) = 0.86, p =
0.650). The RMSEA (.00 < .05) suggest a satisfactory fit. The PNFI (.20 < .50)
suggests a poor fit., The model is not significantly different from a baseline
model (Chi2(2) = 0.86, p = 0.650). The SRMR (.02 < .08) suggest a satisfactory
fit. The PNFI (.20 < .50) suggests a poor fit., The model is not significantly
different from a baseline model (Chi2(2) = 0.86, p = 0.650). The RFI (.97 >
.90) suggest a satisfactory fit. The PNFI (.20 < .50) suggests a poor fit. and
The model is not significantly different from a baseline model (Chi2(2) = 0.86,
p = 0.650). The IFI (.01 > .90) suggest a satisfactory fit. The PNFI (.20 <
.50) suggests a poor fit.Run SEM
parameters::model_parameters(fit, standardize = TRUE,
component = c("regression", "defined")) %>%
print_html()| Model Summary | |||||
|---|---|---|---|---|---|
| Parameter | Coefficient | SE | 95% CI | z | p |
| Regression | |||||
| intent.to.apply ~ perceived.value (a) | 0.81 | 0.10 | (0.62, 1.00) | 8.28 | < .001 |
| intent.to.apply ~ external.pressure (b) | 0.09 | 0.11 | (-0.12, 0.31) | 0.86 | 0.390 |
| intent.to.apply ~ perceived.control (c) | -0.13 | 0.12 | (-0.36, 0.10) | -1.07 | 0.282 |
| application.behaviour ~ intent.to.apply (d) | 0.35 | 0.12 | (0.12, 0.58) | 3.00 | 0.003 |
| application.behaviour ~ perceived.control (e) | 0.34 | 0.12 | (0.10, 0.57) | 2.84 | 0.005 |
| Defined | |||||
| (value.through.intent) | 0.28 | 0.10 | (0.08, 0.48) | 2.74 | 0.006 |
| (pressure.through.intent) | 0.03 | 0.04 | (-0.05, 0.11) | 0.83 | 0.408 |
| (control.through.intent) | -0.04 | 0.05 | (-0.13, 0.04) | -0.98 | 0.329 |
Reporting results
- Make reference to a figure with your hypothesized model and parameter estimates and report model fit
Note
- The hypothesized model was tested with path analysis and the estimated model is depicted in Figure 1. The model appeared to have good fit χ2 (2) = 0.862, p > .650, SRMR = .02, RMSEA= 0, 90% CI [0, 0.20], CFI = 1.
Reporting results
- Describe significance of the paths
Note
- The perceived value of graduate education predicted intentions to apply to grad school, β = 0.81, Z = 7.21, p < 0.01, but intentions to apply to grad school were not significantly predicted by external pressure to go to grad school, β = 0.10, Z = 0.97, p = 0.33, or perceived control over the outcome of graduate admissions, β =-0.13, Z = -1.11, p =.27. However, intention to go to grad school significantly predicted actually applying, β = 0.35, Z = 2.94, p < 0.01, and so did perceived control over the outcome of graduate admissions, β = 0.34, Z = 2.83, p = 0.01. Intent to apply to graduate school mediated the relationship between perceived value of graduate education and actually applying to graduate school, β= 0.68, Z = 2.66, p = 0.01.
Visualize model
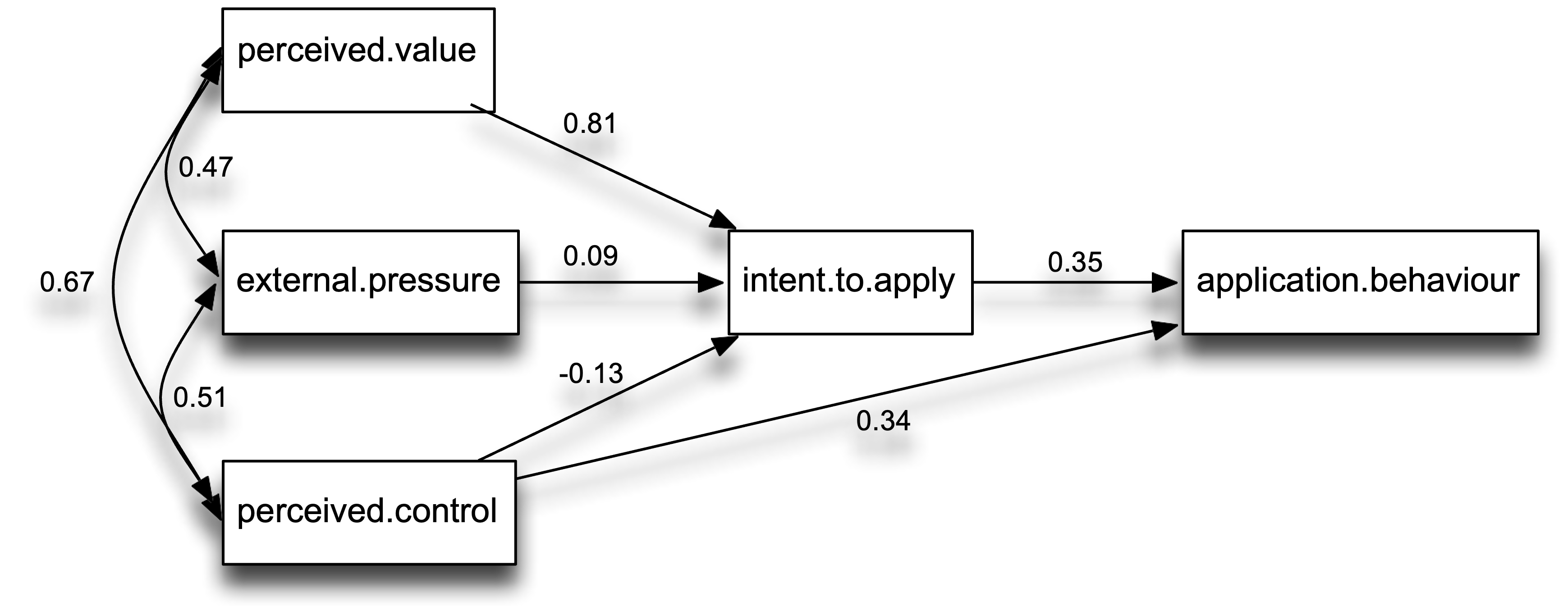
Visualize model
library(semPlot)
# Example of plotting the variables in specific locations
locations = matrix(c(0, 0, .5, 0, -.5, .5, -.5, 0, -.5, -.5), ncol=2, byrow=2)
labels = c("Intent\nTo Apply","Application\nBehaviour","Perceived\nValue","External\nPressure","Perceived\nControl")
diagram = semPaths(fit, whatLabels="std", nodeLabels = labels, layout=locations, sizeMan = 12, rotation=2)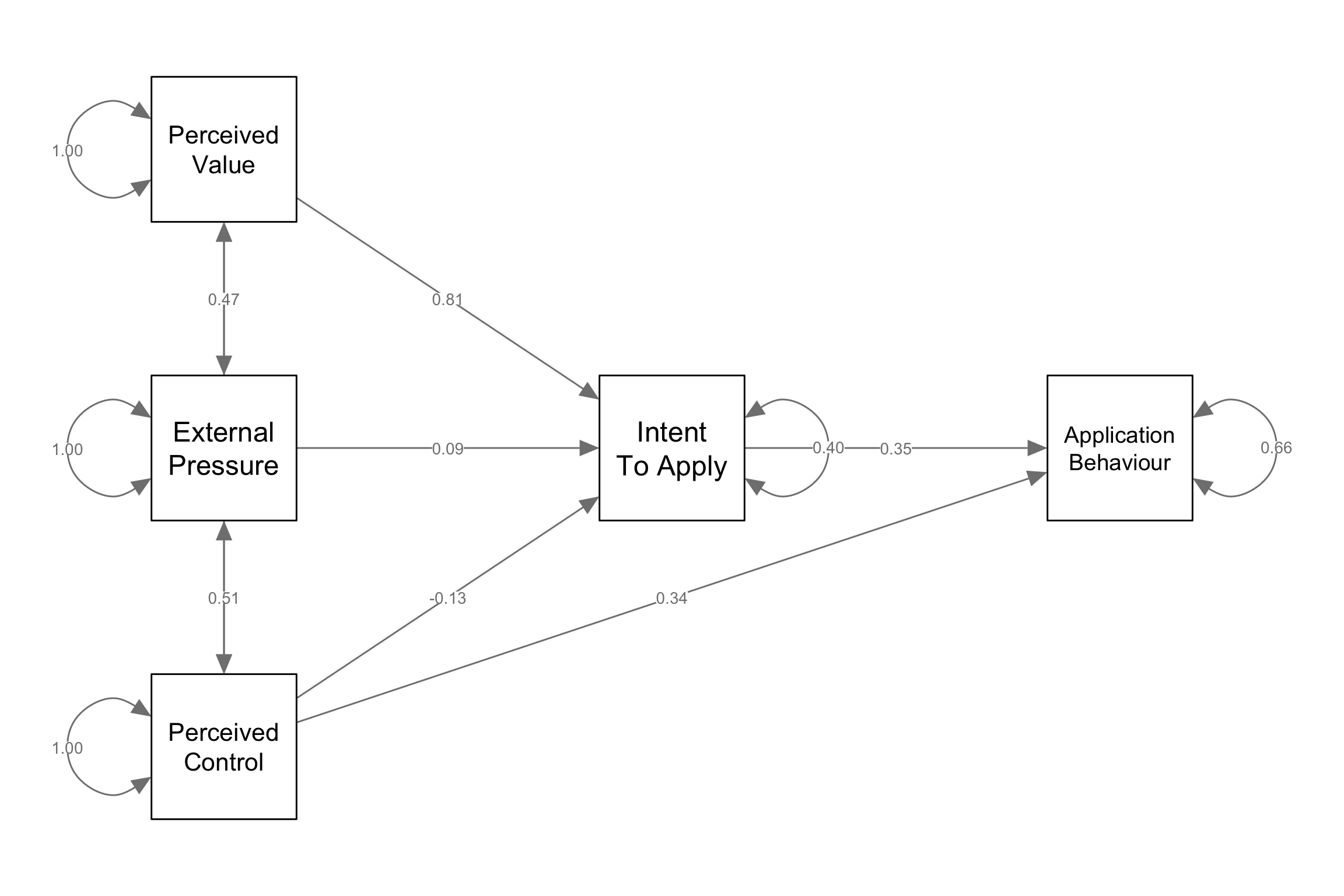
Bad fit
- What if fit incidences are not that good?
Modification (mod) indices
Tell you what the chi-square change would be if you added the path suggested
Can make your model better
- Potential for HARKING!
- Be transparent
Modifications
- Modification indices (look at
micolumn)
| lhs | op | rhs | mi | epc | sepc.lv | sepc.all | sepc.nox |
|---|---|---|---|---|---|---|---|
| intent.to.apply | ~~ | application.behaviour | 0.50 | -4.20 | -4.20 | -0.13 | -0.13 |
| application.behaviour | ~~ | perceived.value | 0.14 | 4.31 | 4.31 | 0.05 | 0.05 |
| application.behaviour | ~~ | external.pressure | 0.43 | 11.74 | 11.74 | 0.07 | 0.07 |
| application.behaviour | ~~ | perceived.control | 0.79 | -15.98 | -15.98 | -0.16 | -0.16 |
| intent.to.apply | ~ | application.behaviour | 0.50 | -0.02 | -0.02 | -0.10 | -0.10 |
| application.behaviour | ~ | perceived.value | 0.35 | 0.28 | 0.28 | 0.12 | 0.12 |
- Anything with \(\chi^2(1)\) > 3.84 is *p* < .05
Best practices
Comparing multiple models
Constraining paths
In SEM you can explicitly test hypotheses about the size of specific paths
Constrain a path to certain value
Constrain two paths to be equal
- Compare model fit of models with constrained and unconstrained paths
Assess alternative hypotheses/models
Constraining paths
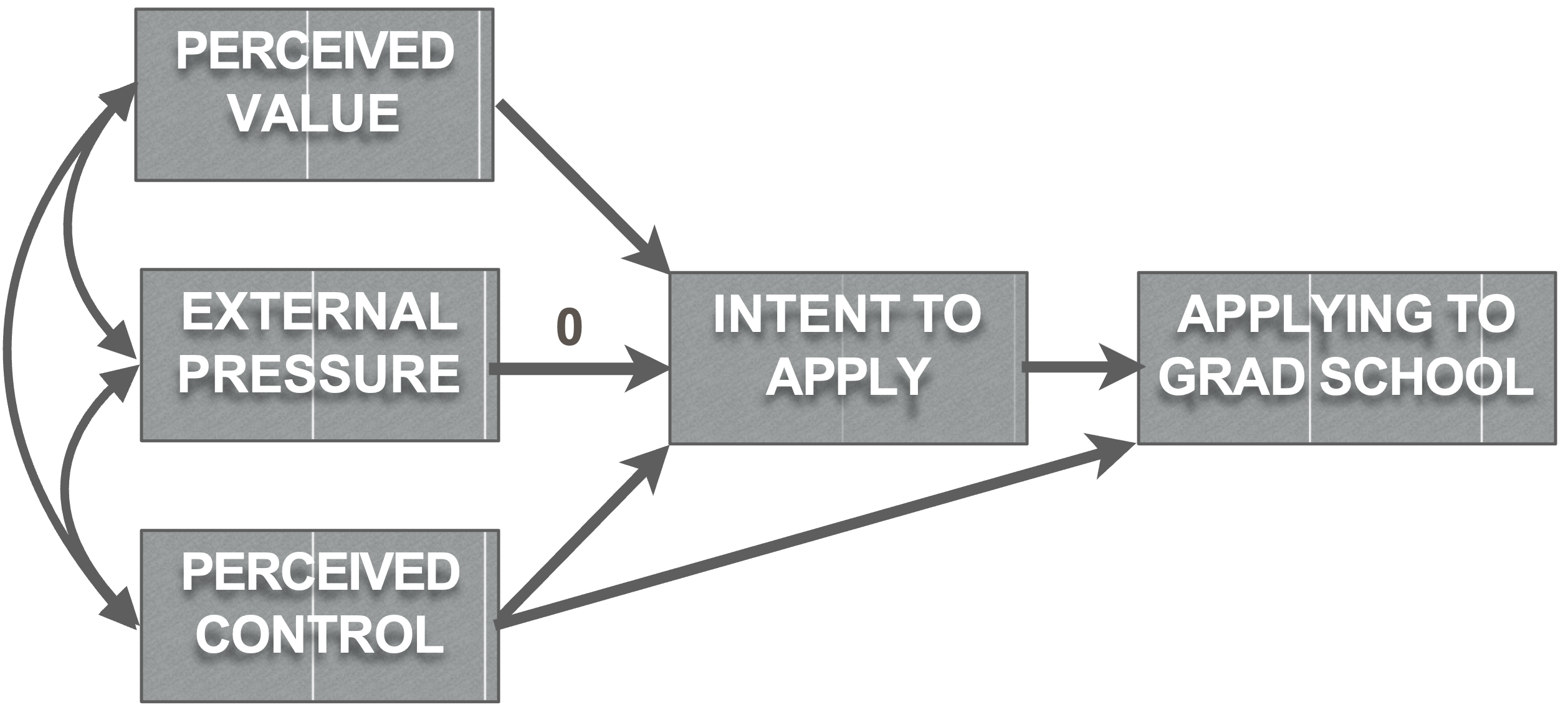
grad_model_constrained = '
intent.to.apply ~ a*perceived.value + 0*external.pressure + c*perceived.control
application.behaviour ~ d*intent.to.apply + perceived.control
perceived.control ~~ perceived.value # These are covariance paths
perceived.control ~~ external.pressure # These are covariance paths
external.pressure ~~ perceived.value # These are covariance paths
value.through.intent:=a*d
control.through.intent:=c*d
'
grad_analysis_constrained =
sem(grad_model_constrained, data=grad, se="bootstrap")Nested model comparisons
If you can create one model from another by the addition or subtraction of parameters, then it is nested
- Model A is said to be nested within Model B, if Model B is a more complicated version of Model A
Evaluating models
Ensure both fit data well
- Report comparative fit indices
- If one sucks, go with least crappy one
- If both suck, don’t compare them
Use LRT test
LRT test
- Test both model fits
| Name | Model | Chi2 | Chi2_df | p_Chi2 | Baseline | Baseline_df | p_Baseline | GFI | AGFI | NFI | NNFI | CFI | RMSEA | RMSEA_CI_low | RMSEA_CI_high | p_RMSEA | RMR | SRMR | RFI | PNFI | IFI | RNI | Loglikelihood | AIC | AIC_wt | BIC | BIC_wt | BIC_adjusted |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fit | lavaan | 0.8615836 | 2 | 0.6499942 | 136.416 | 10 | 0 | 0.9943295 | 0.9574712 | 0.9936841 | 1.045027 | 1 | 0 | 0 | 0.1997409 | 0.6896164 | 3.564326 | 0.0190206 | 0.9684207 | 0.1987368 | 1.008469 | 1.009005 | -993.7334 | 2013.467 | 0.3717899 | 2040.693 | 0.1719716 | 1999.805 |
| grad_analysis_constrained | lavaan | 1.8124921 | 3 | 0.6122203 | 136.416 | 10 | 0 | 0.9886592 | 0.9432959 | 0.9867135 | 1.031312 | 1 | 0 | 0 | 0.1794375 | 0.6671659 | 4.826403 | 0.0301489 | 0.9557116 | 0.2960140 | 1.008901 | 1.009394 | -994.2089 | 2012.418 | 0.6282101 | 2037.550 | 0.8280284 | 1999.807 |
- \(\Delta\) \(\chi^2\) (note: Constraining a path adds 1 df (the model with more DFs is more parsimonious)
Non-nested model comparisons
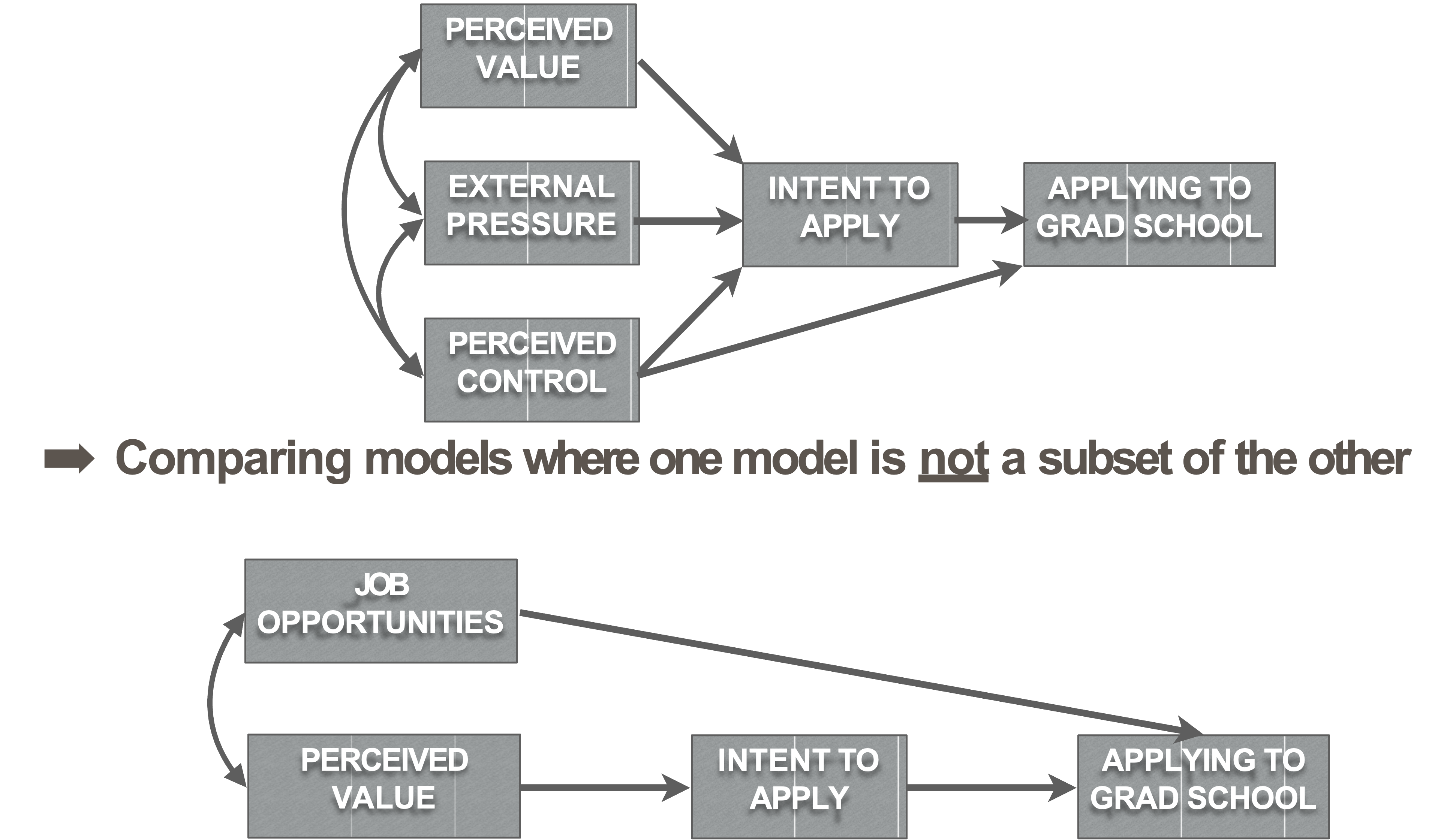
Write-up: Nested models
We wanted to see if the data fit Ajzen’s (1985) Theory of planned behavior (“Unconstrained Model,” Figure 1) better than a constrained model that posits no relationship between external pressure and intention to apply to graduate school (“Constrained Model,” Figure 2). The constrained model fit the data well, SRMR = .03, RMSEA = 0, 90% CI [0, 0.18], CFI = 1, AIC = 2000.42, BIC = 2012.98. A Likelihood Ratio test of the two models suggested that the models fit the data equally well, \(\chi^2\) (1) = 0.95, p = 0.33. Thus, we trimmed this path in the interest of parsimony.
Write-up: Non-nested
We also compared a non-nested model that considered the strongest pathway of our originally hypothesized model in the context of job opportunities (“Opportunities Model,” Figure 3). The opportunities model had good absolute and relative goodness of fit but the relative badness of fit was poor, SRMR = .05, RMSEA =0.28, 90% CI [0.13, 0.44], CFI = 0.96, AIC = 1502.77, BIC = 1519.52. Comparing the Opportunities Model to the Hypothesized Model (Figure 1) using BIC (Kass & Raftery, 1995) reveals that the evidence strongly favors the Opportunities Model, \(BIC_{Hypothesized}\) = 2040.69, ΔBIC= 521.
Practical issues
Sample size: for parameter estimates to be accurate, you should have large samples
How many? Hard to say, but often hundreds are necessary
- http://web.pdx.edu/~newsomj/semclass/ho_sample%20size.pdf
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4334479/
Check out Greg Hancock’s talk https://drive.google.com/file/d/1U6UyYZCJ0sW1muKQZ-VNn9EIBtGWH_uS/view?usp=sharing
- Provides a simple way to calculate power/sample size
Practical issues
- Sample Size: The N:q rule
- Number of people, N
- q number of estimated parameters
- You want the N:q ratio to be 20:1 or greater in a perfect world, 10:1 if you can manage it, at the bare minimum 5:1.
Assumptions
- All assumptions for linear models apply to SEM
- There are robust estimators one can use
- Sattorra-Bentler
sem(model, test="satorra.bentler")
- Sattorra-Bentler
SEM: Summary
SEM is a statistical technique that combines path analysis and factor analysis to allow for statistical modeling of the relationships between observed variables and latent constructs
It focuses heavily on model fit to evaluate how well the proposed model fits the data
It’s a very flexible analysis that has several variations, extensions, and applications
Announcements
Lab
No lab on Wednesday
- We will incorporate SEM into factor analysis lab next week
Next week
- Factor analysis (Aditi and Brooke)
Free SEM workshop
May 8th-10th
Acknowledgments
- Thanks to Elizabeth Page-Gould, Chris Groves, and Erin Buchanan for graciously providing some of the content I used here
PSY 504: Advanced Statistics